Diversity of Multidrug Efflux Genes and Phenotypic Evaluation of the In vitro Resistance Dynamics of Clinical Staphylococcus Aureus Isolates Using Methicillin; a Model β-lactam
- PMID: 28839493
- PMCID: PMC5543611
- DOI: 10.2174/1874285801711010132
Diversity of Multidrug Efflux Genes and Phenotypic Evaluation of the In vitro Resistance Dynamics of Clinical Staphylococcus Aureus Isolates Using Methicillin; a Model β-lactam
Abstract
Objectives: Methicillin-resistant Staphylococcus aureus (MRSA) across the world often leave clinicians with little or no choice of treatment options. The multi-drug efflux (MDE) genes are bacterial survival mechanisms responsible for the pumping out of antibiotics and other biocides from the cytoplasm. Whilst effort is being made in the development of antibiotic adjuvants such as efflux pumps inhibitors, information is needed on the diversity of these MDEs in the circulating S. aureus and on the growth dynamics of the clinical isolates in response to antibiotics is not regularly examined.
Methods: Here, we evaluated the diversity of MDEs in cinical S. aureus recovered in a tertiary academic hospital, Pretoria, South African hospital using PCR and also employed visual minimum inhibitory concentration and quantitative analysis of spectrophometric measurements of bacterial growth in the presence of a model β lactam antibiotic (methicillin), to phenotypically elucidate the resistance pattern of these isolates in response to methicillin.
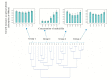
Results: Three major distribution patterns of MDEs were observed in the clinical isolates evaluated. Moreover, norA, nor B and tet38 were present in 98.9% of the isolates while other MDE were present in different proportions ranging from 40 to 98.6% of the isolates. In addition, S. aureus isolates, be it of MRSA or MSSA genotype did not habour the same set of MDEs despite being recovered from the same hospital setting. Finally, we showed that MSSA displayed phenotypic resistance to methicilllin despite the non-detection of the mecA resistance gene.
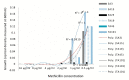
Conclusions: Our data suggest that the growth of S. aureus may be enhanced by β lactams (methicillin) and that MSSA may also display resistance to methicillin and perhaps other β lactam antibiotics. The high prevalence of MDEs suggestive of resistance to a broad spectrum of biocides and fluoroquinolones are particularly disturbing.
Keywords: Methicillin-resistant Staphylococcus aureus (MRSA); Multi-drug efflux (MDE) genes; Quantitative analysis; antibiotic adjuvants; β lactam antibiotic.
Figures

References
-
- Mlynarczyk A., Mlynarczyk G., Jeljaszewicz J. The genome of Staphylococcus aureus: A review. Zentralbl. Bakteriol. 1998;287:277–314. - PubMed
-
- Huet A.A., Raygada J.L., Mendiratta K., Seo S.M., Kaatz G.W. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiol-Sgm. 2008;154:3144–3153. - PubMed
-
- Makgotlho P.E., Kock M.M., Hoosen A., Lekalakala R., Omar S., Dove M., Ehlers M.M. Molecular identification and genotyping of MRSA isolates. FEMS Immunol. Med. Microbiol. 2009;57:104–115. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
