Roles of miR319 and TCP Transcription Factors in Leaf Development
- PMID: 28842549
- PMCID: PMC5619901
- DOI: 10.1104/pp.17.00732
Roles of miR319 and TCP Transcription Factors in Leaf Development
Abstract
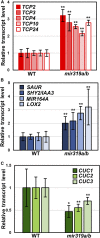
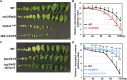
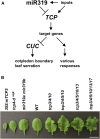
Sophisticated regulation of gene expression, including microRNAs (miRNAs) and their target genes, is required for leaf differentiation, growth, and senescence. The impact of miR319 and its target TEOSINTE BRANCHED1, CYCLOIDEA, and PROLIFERATING CELL NUCLEAR ANTIGEN BINDING FACTOR (TCP) genes on leaf development has been extensively investigated, but the redundancies of these gene families often interfere with the evaluation of their function and regulation in the developmental context. Here, we present the genetic evidence of the involvement of the MIR319 and TCP gene families in Arabidopsis (Arabidopsis thaliana) leaf development. Single mutations in MIR319A and MIR319B genes moderately inhibited the formation of leaf serrations, whereas double mutations increased the extent of this inhibition and resulted in the formation of smooth leaves. Mutations in MIR319 and gain-of-function mutations in the TCP4 gene conferred resistance against miR319 and impaired the cotyledon boundary and leaf serration formation. These mutations functionally associated with CUP-SHAPED COTYLEDON genes, which regulate the cotyledon boundary and leaf serration formation. In contrast, loss-of-function mutations in miR319-targeted and nontargeted TCP genes cooperatively induced the formation of serrated leaves in addition to changes in the levels of their downstream gene transcript. Taken together, these findings demonstrate that the MIR319 and TCP gene families underlie robust and multilayer control of leaf development. This study also provides a framework toward future researches on redundant miRNAs and transcription factors in Arabidopsis and crop plants.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures





References
-
- Aida M, Tasaka M (2006) Genetic control of shoot organ boundaries. Curr Opin Plant Biol 9: 72–77 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Ballester P, Navarrete-Gómez M, Carbonero P, Oñate-Sánchez L, Ferrándiz C (2015) Leaf expansion in Arabidopsis is controlled by a TCP-NGA regulatory module likely conserved in distantly related species. Physiol Plant 155: 21–32 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

