Bacterial Community Structure in the Asian Rice Gall Midge Reveals a Varied Microbiome Rich in Proteobacteria
- PMID: 28842593
- PMCID: PMC5573367
- DOI: 10.1038/s41598-017-09791-0
Bacterial Community Structure in the Asian Rice Gall Midge Reveals a Varied Microbiome Rich in Proteobacteria
Abstract
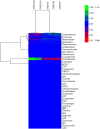
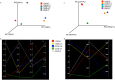
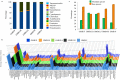
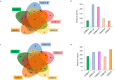
The Asian rice gall midge (ARGM) has emerged as a model gall forming pest of rice. The ARGM infestation of rice results in failure of panicle formation and economic loss. Understanding the molecular basis of ARGM-rice interactions is very crucial in order to control this devastating pest of rice. The current investigation was devised to identify bacterial communities present in the ARGM and in addition the bacterial diversity in the maggots during their interaction with susceptible or resistant rice varieties. Sequencing of 16S rRNA bacterial gene (V3-V4 region) revealed differences in the microflora of the ARGM maggots feeding on susceptible or resistant rice hosts. Results revealed that Wolbachia was the predominant bacterium in pupae and adults while Pseudomonas was predominant in maggots. Further, we observed that members of proteobacteria were predominant across all the samples. There was high species diversity in maggots isolated from susceptible rice and a high representation of unclassified bacteria in maggots isolated from resistant rice. This is the first study that reports variation of microbiome of the ARGM, based on host phenotype from which it was isolated, and results suggest that these variations could have an important role in host's susceptibility.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Infestation of Rice by Gall Midge Influences Density and Diversity of Pseudomonas and Wolbachia in the Host Plant Microbiome.Curr Genomics. 2022 Jun 10;23(2):126-136. doi: 10.2174/1389202923666220401101604. Curr Genomics. 2022. PMID: 36778977 Free PMC article.
-
Feeding on resistant rice leads to enhanced expression of defender against apoptotic cell death (OoDAD1) in the Asian rice gall midge.BMC Plant Biol. 2015 Oct 1;15:235. doi: 10.1186/s12870-015-0618-y. BMC Plant Biol. 2015. PMID: 26428861 Free PMC article.
-
Compatible interaction with its rice host leads to enhanced expression of the gamma subunit of oligosaccharyl transferase in the Asian rice gall midge, Orseolia oryzae.Insect Mol Biol. 2011 Oct;20(5):567-75. doi: 10.1111/j.1365-2583.2011.01085.x. Epub 2011 Jun 13. Insect Mol Biol. 2011. PMID: 21668550
-
A comparative study of microbial community and dynamics of Asaia in the brown planthopper from susceptible and resistant rice varieties.BMC Microbiol. 2019 Jun 24;19(1):139. doi: 10.1186/s12866-019-1512-9. BMC Microbiol. 2019. PMID: 31234788 Free PMC article.
-
Insights into the human oral microbiome.Arch Microbiol. 2018 May;200(4):525-540. doi: 10.1007/s00203-018-1505-3. Epub 2018 Mar 23. Arch Microbiol. 2018. PMID: 29572583 Review.
Cited by
-
Bugs in Bugs: The Role of Probiotics and Prebiotics in Maintenance of Health in Mass-Reared Insects.Insects. 2022 Apr 11;13(4):376. doi: 10.3390/insects13040376. Insects. 2022. PMID: 35447818 Free PMC article. Review.
-
Comparative gut microbiome research through the lens of ecology: theoretical considerations and best practices.Biol Rev Camb Philos Soc. 2025 Apr;100(2):748-763. doi: 10.1111/brv.13161. Epub 2024 Nov 12. Biol Rev Camb Philos Soc. 2025. PMID: 39530277 Free PMC article. Review.
-
Infestation of Rice by Gall Midge Influences Density and Diversity of Pseudomonas and Wolbachia in the Host Plant Microbiome.Curr Genomics. 2022 Jun 10;23(2):126-136. doi: 10.2174/1389202923666220401101604. Curr Genomics. 2022. PMID: 36778977 Free PMC article.
-
Influence of nitric acid on biodegradation of polystyrene and low-density polyethylene by Cephalosporium species.Arch Microbiol. 2022 Jul 15;204(8):489. doi: 10.1007/s00203-022-03089-0. Arch Microbiol. 2022. PMID: 35835894
-
The Diversity of Bacteria Associated with the Invasive Gall Wasp Dryocosmus kuriphilus, Its Galls and a Specialist Parasitoid on Chestnuts.Insects. 2022 Jan 13;13(1):86. doi: 10.3390/insects13010086. Insects. 2022. PMID: 35055929 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

