Structure-based drug discovery for combating influenza virus by targeting the PA-PB1 interaction
- PMID: 28842649
- PMCID: PMC5573363
- DOI: 10.1038/s41598-017-10021-w
Structure-based drug discovery for combating influenza virus by targeting the PA-PB1 interaction
Abstract
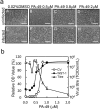
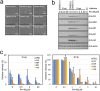
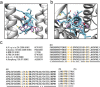
Influenza virus infections are serious public health concerns throughout the world. The development of compounds with novel mechanisms of action is urgently required due to the emergence of viruses with resistance to the currently-approved anti-influenza viral drugs. We performed in silico screening using a structure-based drug discovery algorithm called Nagasaki University Docking Engine (NUDE), which is optimised for a GPU-based supercomputer (DEstination for Gpu Intensive MAchine; DEGIMA), by targeting influenza viral PA protein. The compounds selected by NUDE were tested for anti-influenza virus activity using a cell-based assay. The most potent compound, designated as PA-49, is a medium-sized quinolinone derivative bearing a tetrazole moiety, and it inhibited the replication of influenza virus A/WSN/33 at a half maximal inhibitory concentration of 0.47 μM. PA-49 has the ability to bind PA and its anti-influenza activity was promising against various influenza strains, including a clinical isolate of A(H1N1)pdm09 and type B viruses. The docking simulation suggested that PA-49 interrupts the PA-PB1 interface where important amino acids are mostly conserved in the virus strains tested, suggesting the strain independent utility. Because our NUDE/DEGIMA system is rapid and efficient, it may help effective drug discovery against the influenza virus and other emerging viruses.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Identification of small molecule inhibitors for influenza a virus using in silico and in vitro approaches.PLoS One. 2017 Mar 8;12(3):e0173582. doi: 10.1371/journal.pone.0173582. eCollection 2017. PLoS One. 2017. PMID: 28273150 Free PMC article.
-
Influenza A virus polymerase: an attractive target for next-generation anti-influenza therapeutics.Drug Discov Today. 2018 Mar;23(3):503-518. doi: 10.1016/j.drudis.2018.01.028. Epub 2018 Jan 12. Drug Discov Today. 2018. PMID: 29339107 Review.
-
A Broad Anti-influenza Hybrid Small Molecule That Potently Disrupts the Interaction of Polymerase Acidic Protein-Basic Protein 1 (PA-PB1) Subunits.J Med Chem. 2015 May 14;58(9):3830-42. doi: 10.1021/acs.jmedchem.5b00012. Epub 2015 Apr 20. J Med Chem. 2015. PMID: 25856229
-
Potent and broad-spectrum cycloheptathiophene-3-carboxamide compounds that target the PA-PB1 interaction of influenza virus RNA polymerase and possess a high barrier to drug resistance.Antiviral Res. 2019 May;165:55-64. doi: 10.1016/j.antiviral.2019.03.003. Epub 2019 Mar 15. Antiviral Res. 2019. PMID: 30885750
-
Focusing on the Influenza Virus Polymerase Complex: Recent Progress in Drug Discovery and Assay Development.Curr Med Chem. 2019;26(13):2243-2263. doi: 10.2174/0929867325666180706112940. Curr Med Chem. 2019. PMID: 29984646 Free PMC article. Review.
Cited by
-
Nanobodies: a new frontier in influenza virus neutralization.Folia Microbiol (Praha). 2025 Jul 23. doi: 10.1007/s12223-025-01303-2. Online ahead of print. Folia Microbiol (Praha). 2025. PMID: 40699443 Review.
-
Novel Compounds Identified by Structure-Based Prion Disease Drug Discovery Using In Silico Screening Delay the Progression of an Illness in Prion-Infected Mice.Neurotherapeutics. 2020 Oct;17(4):1836-1849. doi: 10.1007/s13311-020-00903-9. Neurotherapeutics. 2020. PMID: 32767031 Free PMC article.
-
Multi-conformation representation of Mpro identifies promising candidates for drug repurposing against COVID-19.J Mol Model. 2021 Apr 17;27(5):128. doi: 10.1007/s00894-021-04732-1. J Mol Model. 2021. PMID: 33864532 Free PMC article.
-
A Quinolinone Compound Inhibiting the Oligomerization of Nucleoprotein of Influenza A Virus Prevents the Selection of Escape Mutants.Viruses. 2020 Mar 19;12(3):337. doi: 10.3390/v12030337. Viruses. 2020. PMID: 32204549 Free PMC article.
-
PA and PA-X: two key proteins from segment 3 of the influenza viruses.Front Cell Infect Microbiol. 2025 Mar 14;15:1560250. doi: 10.3389/fcimb.2025.1560250. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40160474 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

