EnSVMB: Metagenomics Fragments Classification using Ensemble SVM and BLAST
- PMID: 28842700
- PMCID: PMC5573435
- DOI: 10.1038/s41598-017-09947-y
EnSVMB: Metagenomics Fragments Classification using Ensemble SVM and BLAST
Abstract
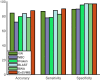
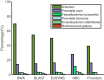
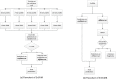
Metagenomics brings in new discoveries and insights into the uncultured microbial world. One fundamental task in metagenomics analysis is to determine the taxonomy of raw sequence fragments. Modern sequencing technologies produce relatively short fragments and greatly increase the number of fragments, and thus make the taxonomic classification considerably more difficult than before. Therefore, fast and accurate techniques are called to classify large-scale fragments. We propose EnSVM (Ensemble Support Vector Machine) and its advanced method called EnSVMB (EnSVM with BLAST) to accurately classify fragments. EnSVM divides fragments into a large confident (or small diffident) set, based on whether the fragments get consistent (or inconsistent) predictions from linear SVMs trained with different k-mers. Empirical study shows that sensitivity and specificity of EnSVM on confident set are higher than 90% and 97%, but on diffident set are lower than 60% and 75%. To further improve the performance on diffident set, EnSVMB takes advantage of best hits of BLAST to reclassify fragments in that set. Experimental results show EnSVM can efficiently and effectively divide fragments into confident and diffident sets, and EnSVMB achieves higher accuracy, sensitivity and more true positives than related state-of-the-art methods and holds comparable specificity with the best of them.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

