Inhibition of the DevSR Two-Component System by Overexpression of Mycobacterium tuberculosis PknB in Mycobacterium smegmatis
- PMID: 28843272
- PMCID: PMC5638771
- DOI: 10.14348/molcells.2017.0076
Inhibition of the DevSR Two-Component System by Overexpression of Mycobacterium tuberculosis PknB in Mycobacterium smegmatis
Abstract
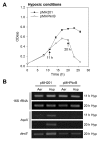
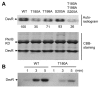
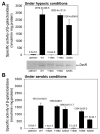
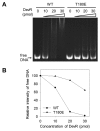
The DevSR (DosSR) two-component system, which is a major regulatory system involved in oxygen sensing in mycobacteria, plays an important role in hypoxic induction of many genes in mycobacteria. We demonstrated that overexpression of the kinase domain of Mycobacterium tuberculosis (Mtb) PknB inhibited transcriptional activity of the DevR response regulator in Mycobacterium smegmatis and that this inhibitory effect was exerted through phosphorylation of DevR on Thr180 within its DNA-binding domain. Moreover, the purified kinase domain of Mtb PknB significantly phosphorylated RegX3, NarL, KdpE, TrcR, DosR, and MtrA response regulators of Mtb that contain the Thr residues corresponding to Thr180 of DevR in their DNA-binding domains, implying that transcriptional activities of these response regulators might also be inhibited when the kinase domain of PknB is overexpressed.
Keywords: DevSR; Ser/Thr protein kinase; mycobacterium; two-component system.
Figures




References
-
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

