The Challenge of Interpreting Glutamate-Receptor Ion-Channel Structures
- PMID: 28844473
- PMCID: PMC5700243
- DOI: 10.1016/j.bpj.2017.07.028
The Challenge of Interpreting Glutamate-Receptor Ion-Channel Structures
Abstract
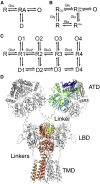
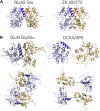
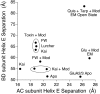
Ion channels activated by glutamate mediate excitatory synaptic transmission in the central nervous system. Similar to other ligand-gated ion channels, their gating cycle begins with transitions from a ligand-free closed state to glutamate-bound active and desensitized states. In an attempt to reveal the molecular mechanisms underlying gating, numerous structures for glutamate receptors have been solved in complexes with agonists, antagonists, allosteric modulators, and auxiliary proteins. The embarrassingly rich library of structures emerging from this work reveals very dynamic molecules with a more complex conformational spectrum than anticipated from functional studies. Unanticipated conformations solved for complexes with competitive antagonists and a lack of understanding of the structural basis for ion channel subconductance states further highlight challenges that have yet to be addressed.
Published by Elsevier Inc.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

