Drosophila PAF1 Modulates PIWI/piRNA Silencing Capacity
- PMID: 28844648
- PMCID: PMC5611886
- DOI: 10.1016/j.cub.2017.07.052
Drosophila PAF1 Modulates PIWI/piRNA Silencing Capacity
Abstract
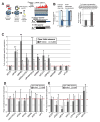
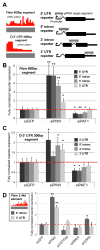
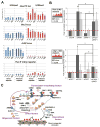
To test the directness of factors in initiating PIWI-directed gene silencing, we employed a Piwi-interacting RNA (piRNA)-targeted reporter assay in Drosophila ovary somatic sheet (OSS) cells [1]. This assay confirmed direct silencing roles for piRNA biogenesis factors and PIWI-associated factors [2-12] but suggested that chromatin-modifying proteins may act downstream of the initial silencing event. Our data also revealed that RNA-polymerase-II-associated proteins like PAF1 and RTF1 antagonize PIWI-directed silencing. PAF1 knockdown enhances PIWI silencing of reporters when piRNAs target the transcript region proximal to the promoter. Loss of PAF1 suppresses endogenous transposable element (TE) transcript maturation, whereas a subset of gene transcripts and long-non-coding RNAs adjacent to TE insertions are affected by PAF1 knockdown in a similar fashion to piRNA-targeted reporters. Additionally, transcription activation at specific TEs and TE-adjacent loci during PIWI knockdown is suppressed when PIWI and PAF1 levels are both reduced. Our study suggests a mechanistic conservation between fission yeast PAF1 repressing AGO1/small interfering RNA (siRNA)-directed silencing [13, 14] and Drosophila PAF1 opposing PIWI/piRNA-directed silencing.
Keywords: Drosophila; PAF1; Piwi; gene silencing; piRNA.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

References
-
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. - PubMed
-
- Iwasaki YW, Murano K, Ishizu H, Shibuya A, Iyoda Y, Siomi MC, Siomi H, Saito K. Piwi Modulates Chromatin Accessibility by Regulating Multiple Factors Including Histone H1 to Repress Transposons. Molecular cell. 2016;63:408–419. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

