Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain
- PMID: 28844694
- PMCID: PMC6791823
- DOI: 10.1016/j.cell.2017.07.036
Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain
Abstract
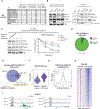
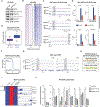
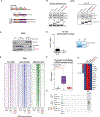
Alterations in transcriptional regulators can orchestrate oncogenic gene expression programs in cancer. Here, we show that the BRG1/BRM-associated factor (BAF) chromatin remodeling complex, which is mutated in over 20% of human tumors, interacts with EWSR1, a member of a family of proteins with prion-like domains (PrLD) that are frequent partners in oncogenic fusions with transcription factors. In Ewing sarcoma, we find that the BAF complex is recruited by the EWS-FLI1 fusion protein to tumor-specific enhancers and contributes to target gene activation. This process is a neomorphic property of EWS-FLI1 compared to wild-type FLI1 and depends on tyrosine residues that are necessary for phase transitions of the EWSR1 prion-like domain. Furthermore, fusion of short fragments of EWSR1 to FLI1 is sufficient to recapitulate BAF complex retargeting and EWS-FLI1 activities. Our studies thus demonstrate that the physical properties of prion-like domains can retarget critical chromatin regulatory complexes to establish and maintain oncogenic gene expression programs.
Keywords: EWS-FLI1; Ewing sarcoma; enhancers; epigenetics; intrinsically disordered proteins; mSWI/SNF (BAF) complexes; microsatellite repeats; phase transition; pioneer factor; prion-like domains.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Prion-like Domains Program Ewing's Sarcoma.Cell. 2017 Sep 21;171(1):30-31. doi: 10.1016/j.cell.2017.09.010. Cell. 2017. PMID: 28938120 Free PMC article.
References
-
- Aguzzi A, and Altmeyer M. (2016). Phase separation: linking cellular compartmentalization to disease. Trends Cell Biol. 26, 547–558. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

