Analysis of deep learning methods for blind protein contact prediction in CASP12
- PMID: 28845538
- PMCID: PMC5871922
- DOI: 10.1002/prot.25377
Analysis of deep learning methods for blind protein contact prediction in CASP12
Abstract
Here we present the results of protein contact prediction achieved in CASP12 by our RaptorX-Contact server, which is an early implementation of our deep learning method for contact prediction. On a set of 38 free-modeling target domains with a median family size of around 58 effective sequences, our server obtained an average top L/5 long- and medium-range contact accuracy of 47% and 44%, respectively (L = length). A complete implementation has an average accuracy of 59% and 57%, respectively. Our deep learning method formulates contact prediction as a pixel-level image labeling problem and simultaneously predicts all residue pairs of a protein using a combination of two deep residual neural networks, taking as input the residue conservation information, predicted secondary structure and solvent accessibility, contact potential, and coevolution information. Our approach differs from existing methods mainly in (1) formulating contact prediction as a pixel-level image labeling problem instead of an image-level classification problem; (2) simultaneously predicting all contacts of an individual protein to make effective use of contact occurrence patterns; and (3) integrating both one-dimensional and two-dimensional deep convolutional neural networks to effectively learn complex sequence-structure relationship including high-order residue correlation. This paper discusses the RaptorX-Contact pipeline, both contact prediction and contact-based folding results, and finally the strength and weakness of our method.
Keywords: CASP; coevolution analysis; deep learning; protein contact prediction; protein folding.
© 2017 Wiley Periodicals, Inc.
Figures
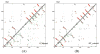

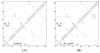


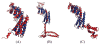

References
-
- de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nature reviews Genetics. 2013;14(4):249–61. - PubMed
-
- Jones DT, et al. PSICOV: precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28(2):184–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

