Rare, high-affinity anti-pathogen antibodies from human repertoires, discovered using microfluidics and molecular genomics
- PMID: 28846502
- PMCID: PMC5680809
- DOI: 10.1080/19420862.2017.1371383
Rare, high-affinity anti-pathogen antibodies from human repertoires, discovered using microfluidics and molecular genomics
Abstract
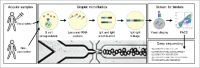
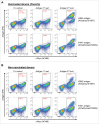
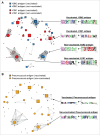
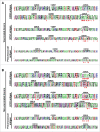
Affinity-matured, functional anti-pathogen antibodies are present at low frequencies in natural human repertoires. These antibodies are often excellent candidates for therapeutic monoclonal antibodies. However, mining natural human antibody repertoires is a challenge. In this study, we demonstrate a new method that uses microfluidics, yeast display, and deep sequencing to identify 247 natively paired anti-pathogen single-chain variable fragments (scFvs), which were initially as rare as 1 in 100,000 in the human repertoires. Influenza A vaccination increased the frequency of influenza A antigen-binding scFv within the peripheral B cell repertoire from <0.1% in non-vaccinated donors to 0.3-0.4% in vaccinated donors, whereas pneumococcus vaccination did not increase the frequency of antigen-binding scFv. However, the pneumococcus scFv binders from the vaccinated library had higher heavy and light chain Replacement/Silent mutation (R/S) ratios, a measure of affinity maturation, than the pneumococcus binders from the corresponding non-vaccinated library. Thus, pneumococcus vaccination may increase the frequency of affinity-matured antibodies in human repertoires. We synthesized 10 anti-influenza A and nine anti-pneumococcus full-length antibodies that were highly abundant among antigen-binding scFv. All 10 anti-influenza A antibodies bound the appropriate antigen at KD<10 nM and neutralized virus in cellular assays. All nine anti-pneumococcus full-length antibodies bound at least one polysaccharide serotype, and 71% of the anti-pneumococcus antibodies that we tested were functional in cell killing assays. Our approach has future application in a variety of fields, including the development of therapeutic antibodies for emerging viral diseases, autoimmune disorders, and cancer.
Keywords: Influenza A; antibody repertoire; deep sequencing; microfluidics; pneumococcus.
Figures

References
-
- Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, Kuzmina N, et al.. Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Cell. 2016;164(3):392-405. doi:10.1016/j.cell.2015.12.022. PMID:26806128 - DOI - PMC - PubMed
-
- Gilchuk I, Gilchuk P, Sapparapu G, Lampley R, Singh V, Kose N, Blum DL, Hughes LJ, Satheshkumar PS, Townsend MB, et al.. Cross-Neutralizing and Protective Human Antibody Specificities to Poxvirus Infections. Cell. 2016;167(3):684-694.e9. doi:10.1016/j.cell.2016.09.049. PMID:27768891 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
