The Sea as a Rich Source of Structurally Unique Glycosaminoglycans and Mimetics
- PMID: 28846656
- PMCID: PMC5620642
- DOI: 10.3390/microorganisms5030051
The Sea as a Rich Source of Structurally Unique Glycosaminoglycans and Mimetics
Abstract
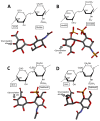
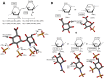
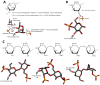
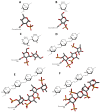
Glycosaminoglycans (GAGs) are sulfated glycans capable of regulating various biological and medical functions. Heparin, heparan sulfate, chondroitin sulfate, dermatan sulfate, keratan sulfate and hyaluronan are the principal classes of GAGs found in animals. Although GAGs are all composed of disaccharide repeating building blocks, the sulfation patterns and the composing alternating monosaccharides vary among classes. Interestingly, GAGs from marine organisms can present structures clearly distinct from terrestrial animals even considering the same class of GAG. The holothurian fucosylated chondroitin sulfate, the dermatan sulfates with distinct sulfation patterns extracted from ascidian species, the sulfated glucuronic acid-containing heparan sulfate isolated from the gastropode Nodipecten nodosum, and the hybrid heparin/heparan sulfate molecule obtained from the shrimp Litopenaeus vannamei are some typical examples. Besides being a rich source of structurally unique GAGs, the sea is also a wealthy environment of GAG-resembling sulfated glycans. Examples of these mimetics are the sulfated fucans and sulfated galactans found in brown, red and green algae, sea urchins and sea cucumbers. For adequate visualization, representations of all discussed molecules are given in both Haworth projections and 3D models.
Keywords: chondroitin sulfate; glycosaminoglycans; heparan sulfate; heparin; sulfated fucans; sulfated galactans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
NMR structural determination of unique invertebrate glycosaminoglycans endowed with medical properties.Carbohydr Res. 2015 Sep 2;413:41-50. doi: 10.1016/j.carres.2015.05.004. Epub 2015 May 27. Carbohydr Res. 2015. PMID: 26083200 Review.
-
Specific sulfation and glycosylation-a structural combination for the anticoagulation of marine carbohydrates.Front Cell Infect Microbiol. 2014 Mar 6;4:33. doi: 10.3389/fcimb.2014.00033. eCollection 2014. Front Cell Infect Microbiol. 2014. PMID: 24639954 Free PMC article. Review.
-
SPR Sensor-Based Analysis of the Inhibition of Marine Sulfated Glycans on Interactions between Monkeypox Virus Proteins and Glycosaminoglycans.Mar Drugs. 2023 Apr 25;21(5):264. doi: 10.3390/md21050264. Mar Drugs. 2023. PMID: 37233458 Free PMC article.
-
Source-induced fragmentation of heparin, heparan, and galactosaminoglycans and application.Anal Chem. 2009 Mar 15;81(6):2332-43. doi: 10.1021/ac802626e. Anal Chem. 2009. PMID: 19228018
-
Holothurian fucosylated chondroitin sulfate.Mar Drugs. 2014 Jan 9;12(1):232-54. doi: 10.3390/md12010232. Mar Drugs. 2014. PMID: 24413804 Free PMC article. Review.
Cited by
-
Extraction, Isolation, Characterization, and Biological Activity of Sulfated Polysaccharides Present in Ascidian Viscera Microcosmus exasperatus.Pharmaceuticals (Basel). 2023 Oct 3;16(10):1401. doi: 10.3390/ph16101401. Pharmaceuticals (Basel). 2023. PMID: 37895872 Free PMC article.
-
Sulfated Polysaccharides from Macroalgae-A Simple Roadmap for Chemical Characterization.Polymers (Basel). 2023 Jan 12;15(2):399. doi: 10.3390/polym15020399. Polymers (Basel). 2023. PMID: 36679279 Free PMC article. Review.
-
Marine Carbohydrate-Based Compounds with Medicinal Properties.Mar Drugs. 2018 Jul 9;16(7):233. doi: 10.3390/md16070233. Mar Drugs. 2018. PMID: 29987239 Free PMC article. Review.
-
Biomaterials from the sea: Future building blocks for biomedical applications.Bioact Mater. 2021 Apr 29;6(12):4255-4285. doi: 10.1016/j.bioactmat.2021.04.028. eCollection 2021 Dec. Bioact Mater. 2021. PMID: 33997505 Free PMC article. Review.
-
Identification and Signature Sequences of Bacterial Δ4,5Hexuronate-2-O-Sulfatases.Front Microbiol. 2019 Apr 5;10:704. doi: 10.3389/fmicb.2019.00704. eCollection 2019. Front Microbiol. 2019. PMID: 31024490 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

