A heterochromatin-dependent transcription machinery drives piRNA expression
- PMID: 28847004
- PMCID: PMC5590728
- DOI: 10.1038/nature23482
A heterochromatin-dependent transcription machinery drives piRNA expression
Abstract
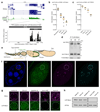
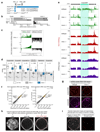
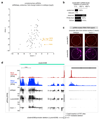
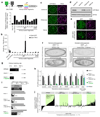
Nuclear small RNA pathways safeguard genome integrity by establishing transcription-repressing heterochromatin at transposable elements. This inevitably also targets the transposon-rich source loci of the small RNAs themselves. How small RNA source loci are efficiently transcribed while transposon promoters are potently silenced is not understood. Here we show that, in Drosophila, transcription of PIWI-interacting RNA (piRNA) clusters-small RNA source loci in animal gonads-is enforced through RNA polymerase II pre-initiation complex formation within repressive heterochromatin. This is accomplished through Moonshiner, a paralogue of a basal transcription factor IIA (TFIIA) subunit, which is recruited to piRNA clusters via the heterochromatin protein-1 variant Rhino. Moonshiner triggers transcription initiation within piRNA clusters by recruiting the TATA-box binding protein (TBP)-related factor TRF2, an animal TFIID core variant. Thus, transcription of heterochromatic small RNA source loci relies on direct recruitment of the core transcriptional machinery to DNA via histone marks rather than sequence motifs, a concept that we argue is a recurring theme in evolution.
Conflict of interest statement
The authors declare no competing financial interests
Figures









Comment in
-
Molecular biology: Rhino gives voice to silent chromatin.Nature. 2017 Sep 6;549(7670):38-39. doi: 10.1038/549038a. Nature. 2017. PMID: 28880287 No abstract available.
-
Chromatin: Probing a piRNA paradox.Nat Rev Genet. 2017 Nov;18(11):638-639. doi: 10.1038/nrg.2017.76. Epub 2017 Sep 11. Nat Rev Genet. 2017. PMID: 28890535 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

