Public antibodies to malaria antigens generated by two LAIR1 insertion modalities
- PMID: 28847005
- PMCID: PMC5635981
- DOI: 10.1038/nature23670
Public antibodies to malaria antigens generated by two LAIR1 insertion modalities
Abstract
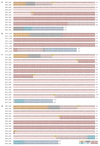
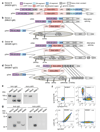
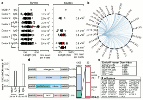
In two previously described donors, the extracellular domain of LAIR1, a collagen-binding inhibitory receptor encoded on chromosome 19 (ref. 1), was inserted between the V and DJ segments of an antibody. This insertion generated, through somatic mutations, broadly reactive antibodies against RIFINs, a type of variant antigen expressed on the surface of Plasmodium falciparum-infected erythrocytes. To investigate how frequently such antibodies are produced in response to malaria infection, we screened plasma from two large cohorts of individuals living in malaria-endemic regions. Here we report that 5-10% of malaria-exposed individuals, but none of the European blood donors tested, have high levels of LAIR1-containing antibodies that dominate the response to infected erythrocytes without conferring enhanced protection against febrile malaria. By analysing the antibody-producing B cell clones at the protein, cDNA and gDNA levels, we characterized additional LAIR1 insertions between the V and DJ segments and discovered a second insertion modality whereby the LAIR1 exon encoding the extracellular domain and flanking intronic sequences are inserted into the switch region. By exon shuffling, this mechanism leads to the production of bispecific antibodies in which the LAIR1 domain is precisely positioned at the elbow between the VH and CH1 domains. Additionally, in one donor the genomic DNA encoding the VH and CH1 domains was deleted, leading to the production of a camel-like LAIR1-containing antibody. Sequencing of the switch regions of memory B cells from European blood donors revealed frequent templated inserts originating from transcribed genes that, in rare cases, comprised exons with orientations and frames compatible with expression. These results reveal different modalities of LAIR1 insertion that lead to public and dominant antibodies against infected erythrocytes and suggest that insertion of templated DNA represents an additional mechanism of antibody diversification that can be selected in the immune response against pathogens and exploited for B cell engineering.
Conflict of interest statement
The authors declare competing financial interests. A.L. is the scientific founder and shareholder of Humabs BioMed. F.S. is a shareholder of Humabs BioMed.
Figures










Comment in
-
Embracing diversity gives antibodies the power to bind.Immunol Cell Biol. 2017 Nov;95(10):862-863. doi: 10.1038/icb.2017.75. Epub 2017 Oct 24. Immunol Cell Biol. 2017. PMID: 29066798 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

