DNA replication through a chromatin environment
- PMID: 28847824
- PMCID: PMC5577465
- DOI: 10.1098/rstb.2016.0287
DNA replication through a chromatin environment
Abstract
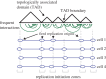
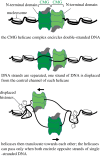
Compaction of the genome into the nuclear space is achieved by wrapping DNA around octameric assemblies of histone proteins to form nucleosomes, the fundamental repeating unit of chromatin. Aside from providing a means by which to fit larger genomes into the cell, chromatinization of DNA is a crucial means by which the cell regulates access to the genome. While the complex role that chromatin plays in gene transcription has been appreciated for a long time, it is now also apparent that crucial aspects of DNA replication are linked to the biology of chromatin. This review will focus on recent advances in our understanding of how the chromatin environment influences key aspects of DNA replication.This article is part of the themed issue 'Chromatin modifiers and remodellers in DNA repair and signalling'.
Keywords: DNA replication; chromatin; nucleosome.
© 2017 The Author(s).
Conflict of interest statement
We declare we have no competing interests.
Figures

References
-
- Jacob F, Brenner S, Cuzin F (eds). 1963. On the regulation of DNA replication in bacteria. Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
