Bacterial PhyA protein-tyrosine phosphatase-like myo-inositol phosphatases in complex with the Ins(1,3,4,5)P4 and Ins(1,4,5)P3 second messengers
- PMID: 28848052
- PMCID: PMC5655508
- DOI: 10.1074/jbc.M117.787853
Bacterial PhyA protein-tyrosine phosphatase-like myo-inositol phosphatases in complex with the Ins(1,3,4,5)P4 and Ins(1,4,5)P3 second messengers
Abstract
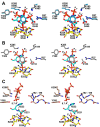
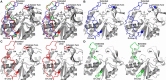
myo-Inositol phosphates (IPs) are important bioactive molecules that have multiple activities within eukaryotic cells, including well-known roles as second messengers and cofactors that help regulate diverse biochemical processes such as transcription and hormone receptor activity. Despite the typical absence of IPs in prokaryotes, many of these organisms express IPases (or phytases) that dephosphorylate IPs. Functionally, these enzymes participate in phosphate-scavenging pathways and in plant pathogenesis. Here, we determined the X-ray crystallographic structures of two catalytically inactive mutants of protein-tyrosine phosphatase-like myo-inositol phosphatases (PTPLPs) from the non-pathogenic bacteria Selenomonas ruminantium (PhyAsr) and Mitsuokella multacida (PhyAmm) in complex with the known eukaryotic second messengers Ins(1,3,4,5)P4 and Ins(1,4,5)P3 Both enzymes bound these less-phosphorylated IPs in a catalytically competent manner, suggesting that IP hydrolysis has a role in plant pathogenesis. The less-phosphorylated IP binding differed in both the myo-inositol ring position and orientation when compared with a previously determined complex structure in the presence of myo-inositol-1,2,3,4,5,6-hexakisphosphate (InsP6 or phytate). Further, we have demonstrated that PhyAsr and PhyAmm have different specificities for Ins(1,2,4,5,6)P5, have identified structural features that account for this difference, and have shown that the absence of these features results in a broad specificity toward Ins(1,2,4,5,6)P5 These features are main-chain conformational differences in loops adjacent to the active site that include the extended loop prior to the penultimate helix, the extended Ω-loop, and a β-hairpin turn of the Phy-specific domain.
Keywords: PTP-like; X-ray crystallography; complex; cysteine phosphatase; inositol 1,4,5-trisphosphate (IP3); inositol phosphate; phosphatase; protein complex; second messenger; substrate specificity.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


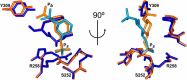

References
-
- Irvine R. F., and Schell M. J. (2001) Back in the water: The return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2, 327–338 - PubMed
-
- York J. D., Odom A. R., Murphy R., Ives E. B., and Wente S. R. (1999) A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285, 96–100 - PubMed
-
- Hanakahi L. A., Bartlet-Jones M., Chappell C., Pappin D., and West S. C. (2000) Binding of inositol phosphate to DNA-PK and stimulation of double-strand break repair. Cell 102, 721–729 - PubMed
-
- Chatterjee S., Sankaranarayanan R., and Sonti R. V. (2003) PhyA, a secreted protein of Xanthomonas oryzae pv. oryzae, is required for optimum virulence and growth on phytic acid as a sole phosphate source. Mol. Plant Microbe Interact. 16, 973–982 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

