Methylomic profiling and replication implicates deregulation of PCSK9 in alcohol use disorder
- PMID: 28848234
- PMCID: PMC5832488
- DOI: 10.1038/mp.2017.168
Methylomic profiling and replication implicates deregulation of PCSK9 in alcohol use disorder
Abstract
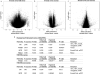
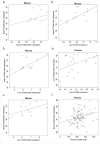
Alcohol use disorder (AUD) is a common and chronic disorder with substantial effects on personal and public health. The underlying pathophysiology is poorly understood but strong evidence suggests significant roles of both genetic and epigenetic components. Given that alcohol affects many organ systems, we performed a cross-tissue and cross-phenotypic analysis of genome-wide methylomic variation in AUD using samples from 3 discovery, 4 replication, and 2 translational cohorts. We identified a differentially methylated region in the promoter of the proprotein convertase subtilisin/kexin 9 (PCSK9) gene that was associated with disease phenotypes. Biological validation showed that PCSK9 promoter methylation is conserved across tissues and positively correlated with expression. Replication in AUD datasets confirmed PCSK9 hypomethylation and a translational mouse model of AUD showed that alcohol exposure leads to PCSK9 downregulation. PCSK9 is primarily expressed in the liver and regulates low-density lipoprotein cholesterol (LDL-C). Our finding of alcohol-induced epigenetic regulation of PCSK9 represents one of the underlying mechanisms between the well-known effects of alcohol on lipid metabolism and cardiovascular risk, with light alcohol use generally being protective while chronic heavy use has detrimental health outcomes.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

