A Vibrio vulnificus VvpM Induces IL-1β Production Coupled with Necrotic Macrophage Death via Distinct Spatial Targeting by ANXA2
- PMID: 28848713
- PMCID: PMC5554522
- DOI: 10.3389/fcimb.2017.00352
A Vibrio vulnificus VvpM Induces IL-1β Production Coupled with Necrotic Macrophage Death via Distinct Spatial Targeting by ANXA2
Abstract
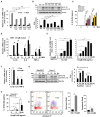
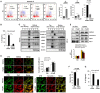
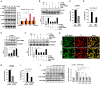
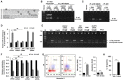
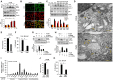
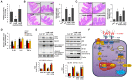
An inflammatory form of phagocyte death evoked by the Gram-negative bacterium Vibrio (V.) vulnificus (WT) is one of hallmarks to promote their colonization, but the virulence factor and infectious mechanism involved in this process remain largely unknown. Here, we identified extracellular metalloprotease VvpM as a new virulence factor and investigated the molecular mechanism of VvpM which acts during the regulation of the inflammatory form of macrophage death and bacterial colonization. Mutation of the vvpM gene appeared to play major role in the prevention of IL-1β production due to V. vulnificus infection in macrophage. However, the recombinant protein (r) VvpM caused IL-1β production coupled with necrotic cell death, which is highly susceptible to the knockdown of annexin A2 (ANXA2) located in both membrane lipid and non-lipid rafts. In lipid rafts, rVvpM recruited NOX enzymes coupled with ANXA2 to facilitate the production of ROS responsible for the epigenetic and transcriptional regulation of NF-κB in the IL-1β promoter. rVvpM acting on non-lipid rafts increased LC3 puncta formation and autophagic flux, which are required for the mRNA expression of Atg5 involved in the autophagosome formation process. The autophagy activation caused by rVvpM induced NLRP3 inflammasome-dependent caspase-1 activation in the promoting of IL-1β production. In mouse models of V. vulnificus infection, the VvpM mutant failed to elevate the level of pro-inflammatory responses closely related to IL-1β production and prevented bacterial colonization. These findings delineate VvpM efficiently regulates two pathogenic pathways that stimulate NF-κB-dependent IL-1β production and autophagy-mediated NLRP3 inflammasome via distinct spatial targeting by ANXA2.
Keywords: ANXA2; Atg5; NLRP3; V. vulnificus; VvpM; cell death; colonization; macrophage.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

