Genome Reduction for Niche Association in Campylobacter Hepaticus, A Cause of Spotty Liver Disease in Poultry
- PMID: 28848714
- PMCID: PMC5554493
- DOI: 10.3389/fcimb.2017.00354
Genome Reduction for Niche Association in Campylobacter Hepaticus, A Cause of Spotty Liver Disease in Poultry
Erratum in
-
Corrigendum: Genome Reduction for Niche Association in Campylobacter Hepaticus, A Cause of Spotty Liver Disease in Poultry.Front Cell Infect Microbiol. 2017 Nov 14;7:480. doi: 10.3389/fcimb.2017.00480. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29159159 Free PMC article.
Abstract
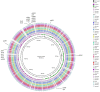
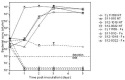
The term "spotty liver disease" (SLD) has been used since the late 1990s for a condition seen in the UK and Australia that primarily affects free range laying hens around peak lay, causing acute mortality and a fall in egg production. A novel thermophilic SLD-associated Campylobacter was reported in the United Kingdom (UK) in 2015. Subsequently, similar isolates occurring in Australia were formally described as a new species, Campylobacter hepaticus. We describe the comparative genomics of 10 C. hepaticus isolates recovered from 5 geographically distinct poultry holdings in the UK between 2010 and 2012. Hierarchical gene-by-gene analyses of the study isolates and representatives of 24 known Campylobacter species indicated that C. hepaticus is most closely related to the major pathogens Campylobacter jejuni and Campylobacter coli. We observed low levels of within-farm variation, even between isolates collected over almost 3 years. With respect to C. hepaticus genome features, we noted that the study isolates had a ~140 Kb reduction in genome size, ~144 fewer genes, and a lower GC content compared to C. jejuni. The most notable reduction was in the subsystem containing genes for iron acquisition and metabolism, supported by reduced growth of C. hepaticus in an iron depletion assay. Genome reduction is common among many pathogens and in C. hepaticus has likely been driven at least in part by specialization following the occupation of a new niche, the chicken liver.
Keywords: Campylobacter hepaticus; genome reduction; niche adaptation; poultry; spotty liver disease.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

