The role of intracellular compartmentalization on tRNA processing and modification
- PMID: 28850002
- PMCID: PMC6103688
- DOI: 10.1080/15476286.2017.1371402
The role of intracellular compartmentalization on tRNA processing and modification
Abstract
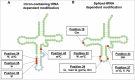
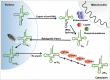
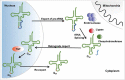
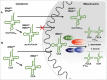
A signature of most eukaryotic cells is the presence of intricate membrane systems. Intracellular organization presumably evolved to provide order, and add layers for regulation of intracellular processes; compartmentalization also forcibly led to the appearance of sophisticated transport systems. With nucleus-encoded tRNAs, it led to the uncoupling of tRNA synthesis from many of the maturation steps it undergoes. It is now clear that tRNAs are actively transported across intracellular membranes and at any point, in any compartment, they can be post-transcriptionally modified; modification enzymes themselves may localize to any of the genome-containing compartments. In the following pages, we describe a number of well-known examples of how intracellular compartmentalization of tRNA processing and modification activities impact the function and fate of tRNAs. We raise the possibility that rates of intracellular transport may influence the level of modification and as such increase the diversity of differentially modified tRNAs in cells.
Keywords: Maturation; modification; nuclear export; retrograde transport; tRNA splicing.
Figures


Similar articles
-
Retrograde nuclear transport from the cytoplasm is required for tRNATyr maturation in T. brucei.RNA Biol. 2018;15(4-5):528-536. doi: 10.1080/15476286.2017.1377878. Epub 2017 Nov 3. RNA Biol. 2018. PMID: 28901827 Free PMC article.
-
The general mRNA exporters Mex67 and Mtr2 play distinct roles in nuclear export of tRNAs in Trypanosoma brucei.Nucleic Acids Res. 2019 Sep 19;47(16):8620-8631. doi: 10.1093/nar/gkz671. Nucleic Acids Res. 2019. PMID: 31392978 Free PMC article.
-
tRNAs in Trypanosoma brucei: genomic organization, expression, and mitochondrial import.Mol Cell Biol. 2002 Jun;22(11):3707-17. doi: 10.1128/MCB.22.11.3707-3716.2002. Mol Cell Biol. 2002. PMID: 11997507 Free PMC article.
-
Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae.Genetics. 2013 May;194(1):43-67. doi: 10.1534/genetics.112.147470. Genetics. 2013. PMID: 23633143 Free PMC article. Review.
-
How the intracellular partitioning of tRNA and tRNA modification enzymes affects mitochondrial function.IUBMB Life. 2018 Dec;70(12):1207-1213. doi: 10.1002/iub.1957. Epub 2018 Oct 25. IUBMB Life. 2018. PMID: 30358065 Free PMC article. Review.
Cited by
-
Transfer RNA function and evolution.RNA Biol. 2018;15(4-5):423-426. doi: 10.1080/15476286.2018.1478942. RNA Biol. 2018. PMID: 30099966 Free PMC article. No abstract available.
-
Pathways to disease from natural variations in human cytoplasmic tRNAs.J Biol Chem. 2019 Apr 5;294(14):5294-5308. doi: 10.1074/jbc.REV118.002982. Epub 2019 Jan 14. J Biol Chem. 2019. PMID: 30643023 Free PMC article. Review.
-
RNA modifications in physiology and disease: towards clinical applications.Nat Rev Genet. 2024 Feb;25(2):104-122. doi: 10.1038/s41576-023-00645-2. Epub 2023 Sep 15. Nat Rev Genet. 2024. PMID: 37714958 Review.
-
tRNA Metabolism and Neurodevelopmental Disorders.Annu Rev Genomics Hum Genet. 2019 Aug 31;20:359-387. doi: 10.1146/annurev-genom-083118-015334. Epub 2019 May 13. Annu Rev Genomics Hum Genet. 2019. PMID: 31082281 Free PMC article. Review.
-
Differentially Expressed tRNA-Derived Small RNAs Co-Sediment Primarily with Non-Polysomal Fractions in Drosophila.Genes (Basel). 2017 Nov 20;8(11):333. doi: 10.3390/genes8110333. Genes (Basel). 2017. PMID: 29156628 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
