A synthetic biology approach for consistent production of plant-made recombinant polyclonal antibodies against snake venom toxins
- PMID: 28850773
- PMCID: PMC5814581
- DOI: 10.1111/pbi.12823
A synthetic biology approach for consistent production of plant-made recombinant polyclonal antibodies against snake venom toxins
Abstract
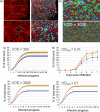
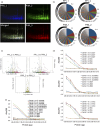
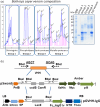
Antivenoms developed from the plasma of hyperimmunized animals are the only effective treatment available against snakebite envenomation but shortage of supply contributes to the high morbidity and mortality toll of this tropical disease. We describe a synthetic biology approach to affordable and cost-effective antivenom production based on plant-made recombinant polyclonal antibodies (termed pluribodies). The strategy takes advantage of virus superinfection exclusion to induce the formation of somatic expression mosaics in agroinfiltrated plants, which enables the expression of complex antibody repertoires in a highly reproducible manner. Pluribodies developed using toxin-binding genetic information captured from peripheral blood lymphocytes of hyperimmunized camels recapitulated the overall binding activity of the immune response. Furthermore, an improved plant-made antivenom (plantivenom) was formulated using an in vitro selected pluribody against Bothrops asper snake venom toxins and has been shown to neutralize a wide range of toxin activities and provide protection against lethal venom doses in mice.
Keywords: molecular pharming; recombinant polyclonal antibodies; snake antivenoms.
© 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures


References
-
- Arnold, C. (2016) Synthetic biology tackles global antivenom shortage. Nature, 532, 292. - PubMed
-
- Bendandi, M. , Marillonnet, S. , Kandzia, R. , Thieme, F. , Nickstadt, A. , Herz, S. , Frode, R. et al. (2010) Rapid, high‐yield production in plants of individualized idiotype vaccines for non‐Hodgkin's lymphoma. Ann. Oncol. 21, 2420–2427. - PubMed
-
- Bolanos, R. (1972) Toxicity of Costa Rican snake venoms for the white mouse. Am. J. Trop. Med. Hyg. 21, 360–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

