The satellite DNA AflaSAT-1 in the A and B chromosomes of the grasshopper Abracris flavolineata
- PMID: 28851268
- PMCID: PMC5575873
- DOI: 10.1186/s12863-017-0548-9
The satellite DNA AflaSAT-1 in the A and B chromosomes of the grasshopper Abracris flavolineata
Abstract
Background: Satellite DNAs (satDNAs) are organized in repetitions directly contiguous to one another, forming long arrays and composing a large portion of eukaryote genomes. These sequences evolve according to the concerted evolution model, and homogenization of repeats is observed at the intragenomic level. Satellite DNAs are the primary component of heterochromatin, located primarily in centromeres and telomeres. Moreover, satDNA enrichment in specific chromosomes has been observed, such as in B chromosomes, that can provide clues about composition, origin and evolution of this chromosome. In this study, we isolated and characterized a satDNA in A and B chromosomes of Abracris flavolineata by integrating cytogenetic, molecular and genomics approaches at intra- and inter-population levels, with the aim to understand the evolution of satDNA and composition of B chromosomes.
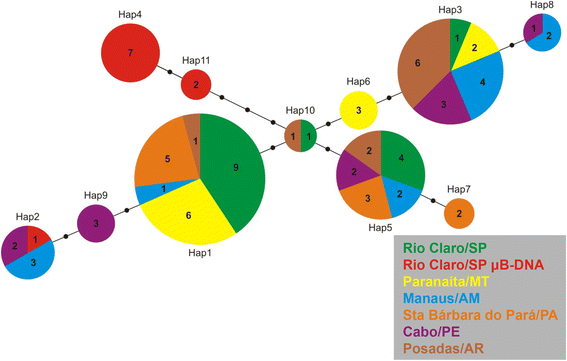
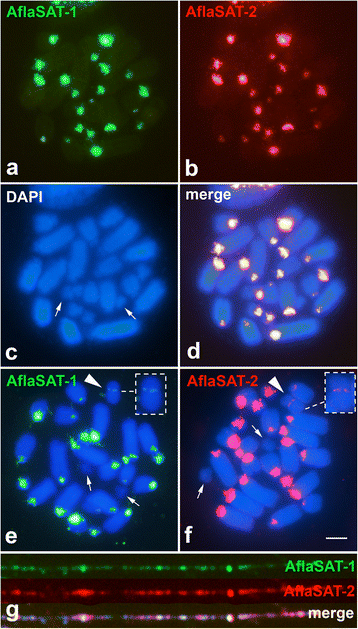
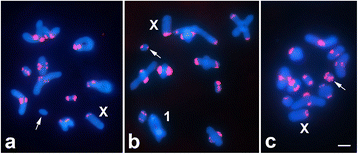
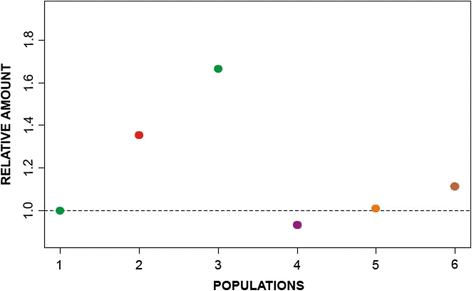
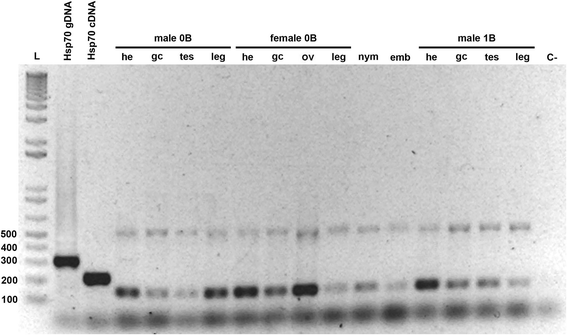
Results: AflaSAT-1 satDNA was shared with other species and in A. flavolineata, was associated with another satDNA, AflaSAT-2. Chromosomal mapping revealed centromeric blocks variable in size in almost all chromosomes (except pair 11) of A complement for both satDNAs, whereas for B chromosome, only a small centromeric signal occurred. In distinct populations, variable number of AflaSAT-1 chromosomal sites correlated with variability in copy number. Instead of such variability, low sequence diversity was observed in A complement, but monomers from B chromosome were more variable, presenting also exclusive mutations. AflaSAT-1 was transcribed in five tissues of adults in distinct life cycle phases.
Conclusions: The sharing of AflaSAT-1 with other species is consistent with the library hypothesis and indicates common origin in a common ancestor; however, AflaSAT-1 was highly amplified in the genome of A. flavolineata. At the population level, homogenization of repeats in distinct populations was documented, but dynamic expansion or elimination of repeats was also observed. Concerning the B chromosome, our data provided new information on the composition in A. flavolineata. Together with previous results, the sequences of heterochromatic nature were not likely highly amplified in the entire B chromosome. Finally, the constitutive transcriptional activity suggests a possible unknown functional role, which should be further investigated.
Keywords: B chromosome; Repetitive DNA; Tandem repeat; Transcription.
Conflict of interest statement
Ethics approval and consent to participate
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Plohl M, Meštrović N, Mravinac B. Satellite DNA evolution. In: Garrido-Ramos MA, editor. Repetitive DNA. Genome dynamics. 7th. Basel: Karger; 2012. pp. 126–152. - PubMed
-
- John B, Miklos GL. Functional aspects of satellite DNA and heterochromatin. In: Bourne GH, Danielli JF, editors. International review of cytology. New York: Academic Press; 1979. pp. 1–114. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

