Comparing microbiota profiles in induced and spontaneous sputum samples in COPD patients
- PMID: 28851370
- PMCID: PMC5576328
- DOI: 10.1186/s12931-017-0645-3
Comparing microbiota profiles in induced and spontaneous sputum samples in COPD patients
Abstract
Background: Induced and spontaneous sputum are used to evaluate the airways microbiota. Whether the sputum types can be used interchangeably in microbiota research is unknown. Our aim was to compare microbiota in induced and spontaneous sputum from COPD patients sampled during the same consultation.
Methods: COPD patients from Bergen, Norway, were followed between 2006/2010, examined during the stable state and exacerbations. 30 patients delivered 36 sample pairs. DNA was extracted by enzymatic and mechanical lysis methods. The V3-V4 region of the 16S rRNA gene was PCR-amplified and prepared for paired-end sequencing. Illumina Miseq System was used for sequencing, and Quantitative Insights Into Microbial Ecology (QIIME) and Stata were used for bioinformatics and statistical analyses.
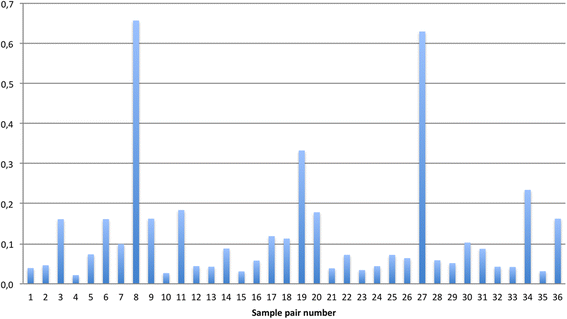
Results: Approximately 4 million sequences were sorted into 1004 different OTUs and further assigned to 106 different taxa. Pair-wise comparison of both taxonomic composition and beta-diversity revealed significant differences in one or both parameters in 1/3 of sample pairs. Alpha-diversity did not differ. Comparing abundances for each taxa identified, showed statistically significant differences between the mean abundances in induced versus spontaneous samples for 15 taxa when disease state was considered. This included potential pathogens like Haemophilus and Moraxella.
Conclusion: When studying microbiota in sputum samples one should take into consideration how samples are collected and avoid the usage of both induced and spontaneous sputum in the same study.
Keywords: COPD; High-throughput sequencing; Microbiota; Sputum.
Conflict of interest statement
Ethics approval and consent to participate
The study was approved by the regional ethical board (REK-Vest Norway), case number 165.08. All participants signed a consent form upon inclusion.
Consent for publication
Not applicable.
Competing interests
ST: Nothing to declare.
MA: Within the last 5 years MA has received a lecture fee from Novartis.
RG: Reports grants from the Norwegian Association of Heart and Lung Patients and EXTRA funds from the Norwegian Foundation for Health and Rehabilitation as well as YaraPraxair during the conduct of the study, grants and personal fees from Boehringer Ingelheim, personal fees from AstraZeneca, and personal fees from GlaxoSmithKline outside the submitted work.
CD: Nothing to declare.
HGW: Nothing to declare.
PSB: Within the last 5 years PSB has received lecture fees or advisory board fees from AstraZeneca, GlaxoSmithKline, Boehringer-Ingelheim, Mundipharma, Chiesi, MSD. PSB is a member of the steering committee and scientific committee of the ECLIPSE study and is PI of the ECLIPSE extension study, both studies sponsored by GlaxoSmithKline.
TME: In the last 5 years TME has received unrestricted grants from Boehringer Ingelheim outside this work, speaker fees from AstraZeneca, Boehringer Ingelheim and GlaxoSmithKline, and support for travel to the AIR conference from InterMune.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Paggiaro PL, Chanez P, Holz O, Ind PW, Djukanovic R, Maestrelli P, Sterk PJ. Sputum induction. Eur Respir J Suppl. 2002;37:3s–8s. - PubMed
-
- Bafadhel M, Haldar K, Barker B, Patel H, Mistry V, Barer MR, Pavord ID, Brightling CE. Airway bacteria measured by quantitative polymerase chain reaction and culture in patients with stable COPD: relationship with neutrophilic airway inflammation, exacerbation frequency, and lung function. Int J Chron Obstruct Pulmon Dis. 2015;10:1075–1083. doi: 10.2147/COPD.S80091. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

