Putrescine independent wound response phenotype is produced by ODC-like RNAi in planarians
- PMID: 28851936
- PMCID: PMC5574924
- DOI: 10.1038/s41598-017-09567-6
Putrescine independent wound response phenotype is produced by ODC-like RNAi in planarians
Abstract
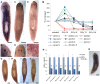
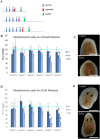
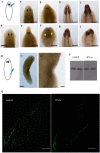
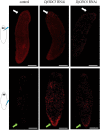
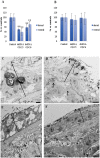
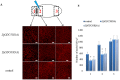
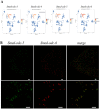
Despite increasing evidence indicates polyamines as a convergence point for signaling pathways, including cell growth and differentiation, a unifying concept to interpret their role is still missing. The activity of ornithine decarboxylase (ODC), the rate-limiting enzyme in polyamine biosynthesis, is tightly regulated by a complex molecular machinery, and the demonstration of the existence of multiple ODC paralogs, lacking decarboxylation activity, suggests additional layers of complexity to the intricate ODC regulatory pathway. Because of their extraordinary regenerative abilities and abundance of stem cells, planarians have potential to contribute to our understanding of polyamine function in an in vivo context. We undertook a study on ODC function in planarians and we found six planarian ODCs (ODC1-6). Five out of six ODC homologs carry substitutions of key aminoacids for enzymatic activity, which makes them theoretically unable to decarboxylate ornithine. Silencing of ODC5 and 6 produced a complex phenotype, by prompting animals to an aberrant response, following chronic injury without tissue removal. Phenotype is neither rescued by putrescine, nor mimicked by difluoromethylornithine treatment. Moreover, the co-silencing of other genes of the ODC regulatory pathway did not modulate phenotype outcome or severity, thus suggesting that the function/s of these ODC-like proteins might be unrelated to decarboxylase activity and putrescine production.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Cohen, S. S. A Guide to Polyamines. Oxford University Press, New York (1998).
-
- Pegg AE. Polyamine metabolism and its importance in neoplastic growth and as a target for chemotherapy. Cancer Res. 1988;48:759–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

