A novel approach for the prediction of species-specific biotransformation of xenobiotic/drug molecules by the human gut microbiota
- PMID: 28852076
- PMCID: PMC5575299
- DOI: 10.1038/s41598-017-10203-6
A novel approach for the prediction of species-specific biotransformation of xenobiotic/drug molecules by the human gut microbiota
Abstract
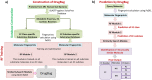
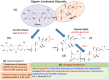
The human gut microbiota is constituted of a diverse group of microbial species harbouring an enormous metabolic potential, which can alter the metabolism of orally administered drugs leading to individual/population-specific differences in drug responses. Considering the large heterogeneous pool of human gut bacteria and their metabolic enzymes, investigation of species-specific contribution to xenobiotic/drug metabolism by experimental studies is a challenging task. Therefore, we have developed a novel computational approach to predict the metabolic enzymes and gut bacterial species, which can potentially carry out the biotransformation of a xenobiotic/drug molecule. A substrate database was constructed for metabolic enzymes from 491 available human gut bacteria. The structural properties (fingerprints) from these substrates were extracted and used for the development of random forest models, which displayed average accuracies of up to 98.61% and 93.25% on cross-validation and blind set, respectively. After the prediction of EC subclass, the specific metabolic enzyme (EC) is identified using a molecular similarity search. The performance was further evaluated on an independent set of FDA-approved drugs and other clinically important molecules. To our knowledge, this is the only available approach implemented as 'DrugBug' tool for the prediction of xenobiotic/drug metabolism by metabolic enzymes of human gut microbiota.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
GutBug: A Tool for Prediction of Human Gut Bacteria Mediated Biotransformation of Biotic and Xenobiotic Molecules Using Machine Learning.J Mol Biol. 2023 Jul 15;435(14):168056. doi: 10.1016/j.jmb.2023.168056. Epub 2023 Mar 22. J Mol Biol. 2023. PMID: 37356904
-
Xenobiotic Metabolism and Gut Microbiomes.PLoS One. 2016 Oct 3;11(10):e0163099. doi: 10.1371/journal.pone.0163099. eCollection 2016. PLoS One. 2016. PMID: 27695034 Free PMC article.
-
HgutMgene-Miner: In silico genome mining tool for deciphering the drug-metabolizing potential of human gut microbiome.Comput Biol Med. 2025 Mar;186:109679. doi: 10.1016/j.compbiomed.2025.109679. Epub 2025 Jan 24. Comput Biol Med. 2025. PMID: 39862468
-
Chemical transformation of xenobiotics by the human gut microbiota.Science. 2017 Jun 23;356(6344):eaag2770. doi: 10.1126/science.aag2770. Science. 2017. PMID: 28642381 Free PMC article. Review.
-
Human Gut Microbiota and Drug Metabolism.Microb Ecol. 2023 Jul;86(1):97-111. doi: 10.1007/s00248-022-02081-x. Epub 2022 Jul 23. Microb Ecol. 2023. PMID: 35869999 Free PMC article. Review.
Cited by
-
Orally Administered Drugs and Their Complicated Relationship with Our Gastrointestinal Tract.Microorganisms. 2024 Jan 24;12(2):242. doi: 10.3390/microorganisms12020242. Microorganisms. 2024. PMID: 38399646 Free PMC article. Review.
-
Computational analysis of the gut microbiota-mediated drug metabolism.Comput Struct Biotechnol J. 2025 Mar 11;27:1472-1481. doi: 10.1016/j.csbj.2025.03.016. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40248646 Free PMC article.
-
Insights from pharmacokinetic models of host-microbiome drug metabolism.Gut Microbes. 2020 May 3;11(3):587-596. doi: 10.1080/19490976.2019.1667724. Epub 2019 Sep 29. Gut Microbes. 2020. PMID: 31564204 Free PMC article.
-
Diet as a Modulator of Intestinal Microbiota in Rheumatoid Arthritis.Nutrients. 2020 Nov 14;12(11):3504. doi: 10.3390/nu12113504. Nutrients. 2020. PMID: 33202579 Free PMC article. Review.
-
Gut microbiome-derived hydrolases-an underrated target of natural product metabolism.Front Cell Infect Microbiol. 2024 Jun 10;14:1392249. doi: 10.3389/fcimb.2024.1392249. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38915922 Free PMC article. Review.
References
-
- Sartor RB, Mazmanian SK. Intestinal microbes in inflammatory bowel diseases. The American Journal of Gastroenterology Supplements. 2012;1:15–21. doi: 10.1038/ajgsup.2012.4. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

