Mapping architectural and transcriptional alterations in non-lesional and lesional epidermis in vitiligo
- PMID: 28852211
- PMCID: PMC5575244
- DOI: 10.1038/s41598-017-10253-w
Mapping architectural and transcriptional alterations in non-lesional and lesional epidermis in vitiligo
Abstract
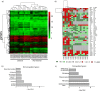
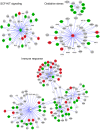
In vitiligo, chronic loss of melanocytes and consequent absence of melanin from the epidermis presents a challenge for long-term tissue maintenance. The stable vitiligo patches are known to attain an irreversible depigmented state. However, the molecular and cellular processes resulting in this remodeled tissue homeostasis is unclear. To investigate the complex interplay of inductive signals and cell intrinsic factors that support the new acquired state, we compared the matched lesional and non-lesional epidermis obtained from stable non-segmental vitiligo subjects. Hierarchical clustering of genome-wide expression of transcripts surprisingly segregated lesional and non-lesional samples in two distinct clades, despite the apparent heterogeneity in the lesions of different vitiligo subjects. Pathway enrichment showed the expected downregulation of melanogenic pathway and a significant downregulation of cornification and keratinocyte differentiation processes. These perturbations could indeed be recapitulated in the lesional epidermal tissue, including blunting of rete-ridges, thickening of stratum corneum and increase in the size of corneocytes. In addition, we identify marked increase in the putrescine levels due to the elevated expression of spermine/spermidine acetyl transferase. Our study provides insights into the intrinsic self-renewing ability of damaged lesional tissue to restore epidermal functionality in vitiligo.
Conflict of interest statement
R.S.G. is the co-founder director on the board of Vyome Biosciences, a biopharmaceutical company in the area of dermatology. K.N. is on the board of Ahammune Biosciences, a biopharmaceutical company in the area of dermatology.
Figures


Similar articles
-
Genome-wide profiling reveals pervasive transcriptional alterations in fibroblasts derived from lesional skin in vitiligo including a reduced potential to proliferate.Exp Dermatol. 2023 Apr;32(4):331-340. doi: 10.1111/exd.14702. Epub 2022 Nov 18. Exp Dermatol. 2023. PMID: 36333875
-
Mechanisms underlying the dysfunction of melanocytes in vitiligo epidermis: role of SCF/KIT protein interactions and the downstream effector, MITF-M.J Pathol. 2004 Apr;202(4):463-75. doi: 10.1002/path.1538. J Pathol. 2004. PMID: 15095274
-
Keratinocyte dysfunction in vitiligo epidermis: cytokine microenvironment and correlation to keratinocyte apoptosis.Histol Histopathol. 2009 Jul;24(7):849-57. doi: 10.14670/HH-24.849. Histol Histopathol. 2009. PMID: 19475531
-
Involvement of non-melanocytic skin cells in vitiligo.Exp Dermatol. 2019 Jun;28(6):667-673. doi: 10.1111/exd.13868. Epub 2019 Feb 11. Exp Dermatol. 2019. PMID: 30582762 Review.
-
Vitiligo.Semin Cutan Med Surg. 1997 Mar;16(1):3-14. doi: 10.1016/s1085-5629(97)80030-2. Semin Cutan Med Surg. 1997. PMID: 9125760 Review.
Cited by
-
Altered epidermal proliferation, differentiation, and lipid composition: Novel key elements in the vitiligo puzzle.Sci Adv. 2022 Sep 2;8(35):eabn9299. doi: 10.1126/sciadv.abn9299. Epub 2022 Sep 2. Sci Adv. 2022. PMID: 36054352 Free PMC article.
-
Micro RNAs upregulated in Vitiligo skin play an important role in its aetiopathogenesis by altering TRP1 expression and keratinocyte-melanocytes cross-talk.Sci Rep. 2019 Jul 12;9(1):10079. doi: 10.1038/s41598-019-46529-6. Sci Rep. 2019. PMID: 31300697 Free PMC article.
-
Development of a Multi-Target Strategy for the Treatment of Vitiligo via Machine Learning and Network Analysis Methods.Front Pharmacol. 2021 Sep 15;12:754175. doi: 10.3389/fphar.2021.754175. eCollection 2021. Front Pharmacol. 2021. PMID: 34603063 Free PMC article.
-
Exploring the influence of gut microbiota metabolites on vitiligo through the gut-skin axis.Front Microbiol. 2025 Jul 9;16:1566267. doi: 10.3389/fmicb.2025.1566267. eCollection 2025. Front Microbiol. 2025. PMID: 40703229 Free PMC article. Review.
-
Transcriptome Analysis and Emerging Driver Identification of CD8+ T Cells in Patients with Vitiligo.Oxid Med Cell Longev. 2019 Nov 26;2019:2503924. doi: 10.1155/2019/2503924. eCollection 2019. Oxid Med Cell Longev. 2019. PMID: 31885781 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

