Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma
- PMID: 28852427
- PMCID: PMC5571486
- DOI: 10.1186/s13148-017-0392-9
Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma
Abstract
Background: DNA methylation has started a recent revolution in genomics biology by identifying key biomarkers for multiple cancers, including oral squamous cell carcinoma (OSCC), the most common head and neck squamous cell carcinoma.
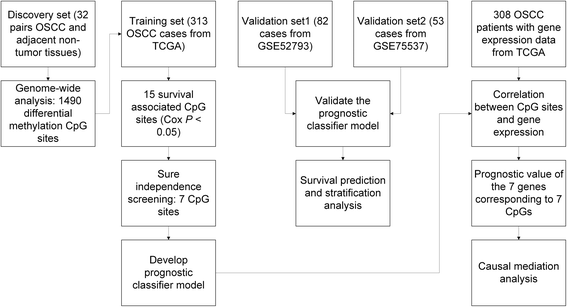
Methods: A multi-stage screening strategy was used to identify DNA-methylation-based signatures for OSCC prognosis. We used The Cancer Genome Atlas (TCGA) data as training set which were validated in two independent datasets from Gene Expression Omnibus (GEO). The correlation between DNA methylation and corresponding gene expression and the prognostic value of the gene expression were explored as well.
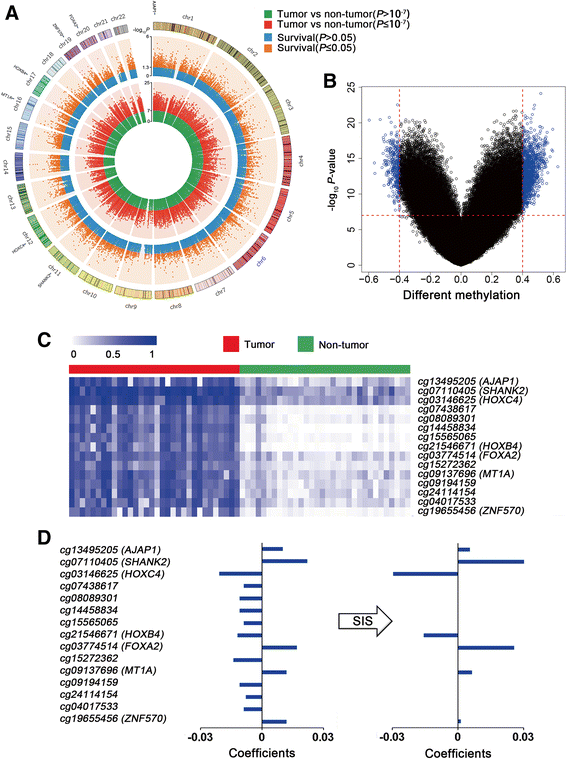
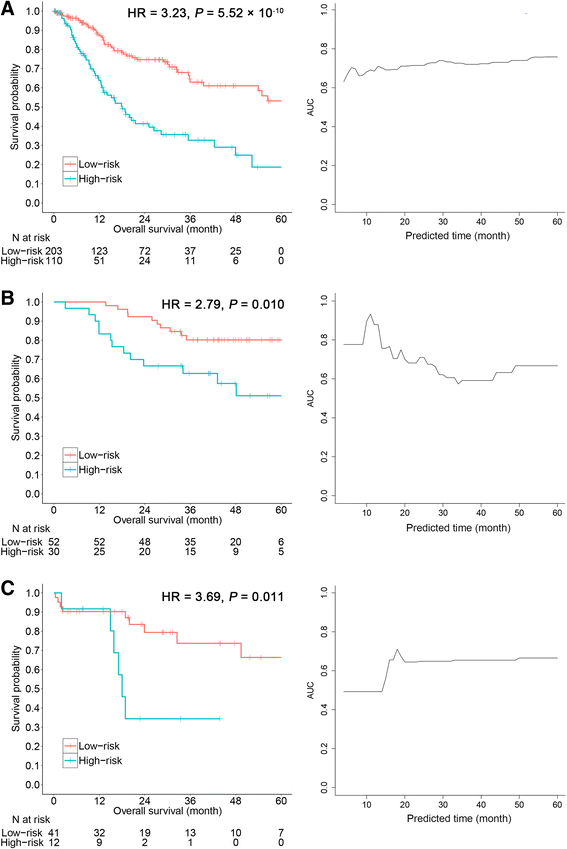
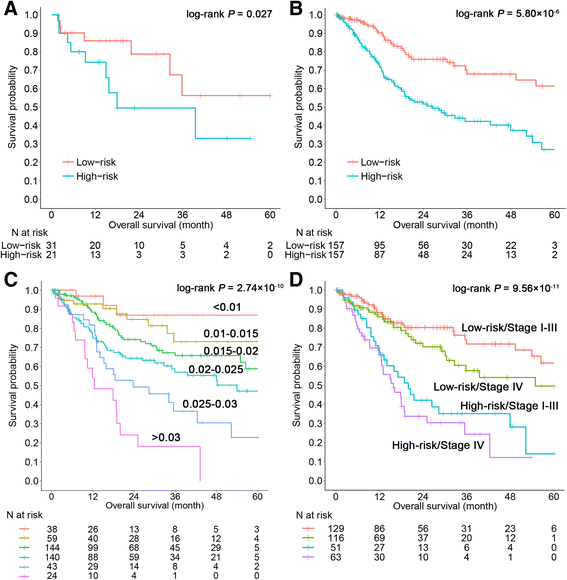
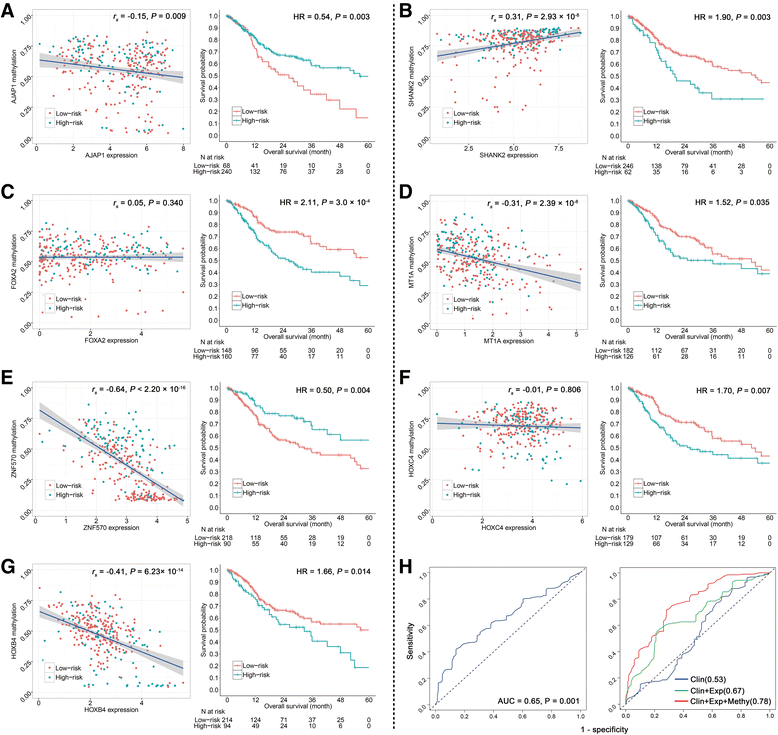
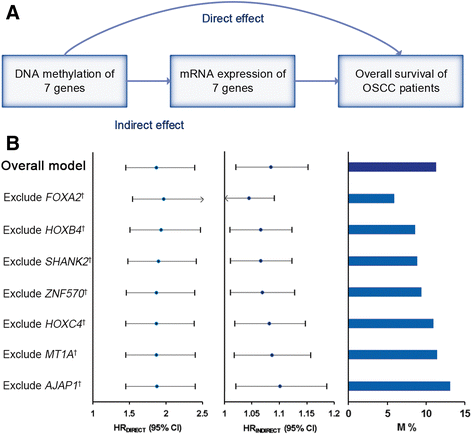
Results: The seven DNA methylation CpG sites were identified which were significantly associated with OSCC overall survival. Prognostic signature, a weighted linear combination of the seven CpG sites, successfully distinguished the overall survival of OSCC patients and had a moderate predictive ability for survival [training set: hazard ratio (HR) = 3.23, P = 5.52 × 10-10, area under the curve (AUC) = 0.76; validation set 1: HR = 2.79, P = 0.010, AUC = 0.67; validation set 2: HR = 3.69, P = 0.011, AUC = 0.66]. Stratification analysis by human papillomavirus status, clinical stage, age, gender, smoking status, and grade retained statistical significance. Expression of genes corresponding to candidate CpG sites (AJAP1, SHANK2, FOXA2, MT1A, ZNF570, HOXC4, and HOXB4) was also significantly associated with patient's survival. Signature integrating of DNA methylation, gene expression, and clinical information showed a superior ability for prognostic prediction (AUC = 0.78).
Conclusion: Prognostic signature integrated of DNA methylation, gene expression, and clinical information provides a better prognostic prediction value for OSCC patients than that with clinical information only.
Keywords: Gene expression; Methylation; Oral squamous cell carcinoma; Overall survival; Prognostic signature.
Conflict of interest statement
Consent for publication
All participants gave written informed consent. All authors have reviewed the manuscript and consented for publication.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

