Complete Genome sequence of the nematicidal Bacillus thuringiensis MYBT18246
- PMID: 28852435
- PMCID: PMC5569534
- DOI: 10.1186/s40793-017-0259-x
Complete Genome sequence of the nematicidal Bacillus thuringiensis MYBT18246
Abstract
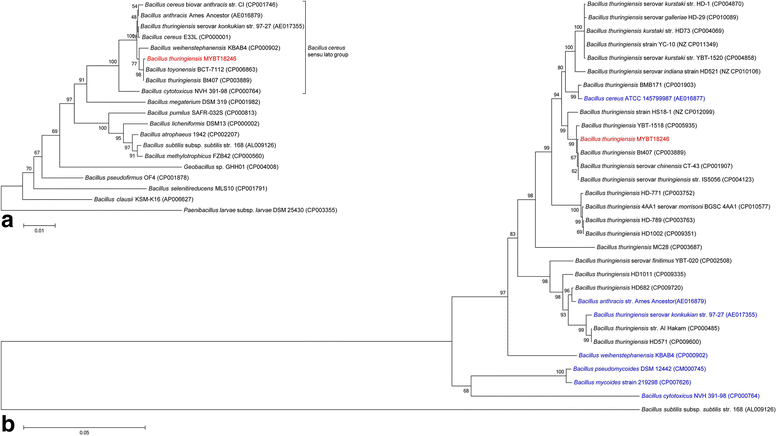
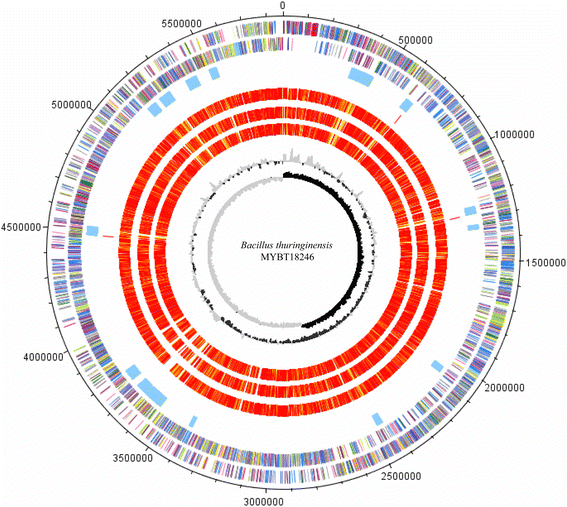
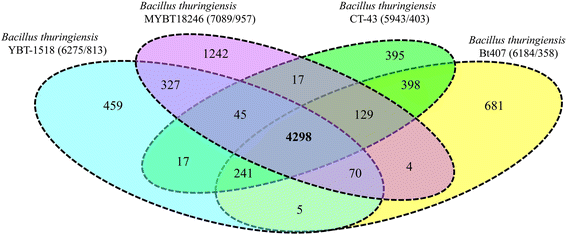
10.1601/nm.5000 is a rod-shaped facultative anaerobic spore forming bacterium of the genus 10.1601/nm.4857. The defining feature of the species is the ability to produce parasporal crystal inclusion bodies, consisting of δ-endotoxins, encoded by cry-genes. Here we present the complete annotated genome sequence of the nematicidal 10.1601/nm.5000 strain MYBT18246. The genome comprises one 5,867,749 bp chromosome and 11 plasmids which vary in size from 6330 bp to 150,790 bp. The chromosome contains 6092 protein-coding and 150 RNA genes, including 36 rRNA genes. The plasmids encode 997 proteins and 4 t-RNA's. Analysis of the genome revealed a large number of mobile elements involved in genome plasticity including 11 plasmids and 16 chromosomal prophages. Three different nematicidal toxin genes were identified and classified according to the Cry toxin naming committee as cry13Aa2, cry13Ba1, and cry13Ab1. Strikingly, these genes are located on the chromosome in close proximity to three separate prophages. Moreover, four putative toxin genes of different toxin classes were identified on the plasmids p120510 (Vip-like toxin), p120416 (Cry-like toxin) and p109822 (two Bin-like toxins). A comparative genome analysis of 10.1601/nm.5000 MYBT18246 with three closely related 10.1601/nm.5000 strains enabled determination of the pan-genome of 10.1601/nm.5000 MYBT18246, revealing a large number of singletons, mostly represented by phage genes, morons and cryptic genes.
Keywords: Bacillus cereus sensu lato; Bacillus thuringiensis; Pan-Core-genome; Parasporal crystal protein; Prophages.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Complete genome sequence of the nematicidal Bacillus thuringiensis MYBT18247.J Biotechnol. 2017 Oct 20;260:48-52. doi: 10.1016/j.jbiotec.2017.09.003. Epub 2017 Sep 9. J Biotechnol. 2017. PMID: 28899808
-
A novel anti-dipteran Bacillus thuringiensis strain: Unusual Cry toxin genes in a highly dynamic plasmid environment.Appl Environ Microbiol. 2021 Mar 1;87(5):e02294-20. doi: 10.1128/AEM.02294-20. Epub 2020 Dec 11. Appl Environ Microbiol. 2021. PMID: 33310715 Free PMC article.
-
Complete genome and plasmid sequence of a novel Bacillus sp. BD59S, a parasporal protein synthesizing bacterium.3 Biotech. 2019 Sep;9(9):318. doi: 10.1007/s13205-019-1849-7. Epub 2019 Aug 3. 3 Biotech. 2019. PMID: 31406640 Free PMC article.
-
The molecular biology of parasporal crystal body formation in Bacillus thuringiensis.Annu Rev Microbiol. 1986;40:549-76. doi: 10.1146/annurev.mi.40.100186.003001. Annu Rev Microbiol. 1986. PMID: 3535652 Review.
-
Recent advancement on chemical arsenal of Bt toxin and its application in pest management system in agricultural field.3 Biotech. 2018 Apr;8(4):201. doi: 10.1007/s13205-018-1223-1. Epub 2018 Mar 29. 3 Biotech. 2018. PMID: 29607282 Free PMC article. Review.
Cited by
-
The Distribution of Several Genomic Virulence Determinants Does Not Corroborate the Established Serotyping Classification of Bacillus thuringiensis.Int J Mol Sci. 2021 Feb 24;22(5):2244. doi: 10.3390/ijms22052244. Int J Mol Sci. 2021. PMID: 33668147 Free PMC article.
-
Genomic Characterization and Probiotic Potency of Bacillus sp. DU-106, a Highly Effective Producer of L-Lactic Acid Isolated From Fermented Yogurt.Front Microbiol. 2018 Sep 20;9:2216. doi: 10.3389/fmicb.2018.02216. eCollection 2018. Front Microbiol. 2018. PMID: 30294310 Free PMC article.
-
Role of plasmid plasticity and mobile genetic elements in the entomopathogen Bacillus thuringiensis serovar israelensis.FEMS Microbiol Rev. 2018 Nov 1;42(6):829-856. doi: 10.1093/femsre/fuy034. FEMS Microbiol Rev. 2018. PMID: 30203090 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

