Identification of Phytochemicals Targeting c-Met Kinase Domain using Consensus Docking and Molecular Dynamics Simulation Studies
- PMID: 28852971
- PMCID: PMC7090793
- DOI: 10.1007/s12013-017-0821-6
Identification of Phytochemicals Targeting c-Met Kinase Domain using Consensus Docking and Molecular Dynamics Simulation Studies
Abstract
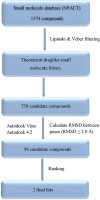
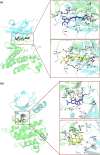
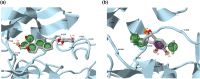
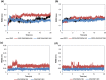
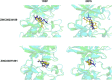
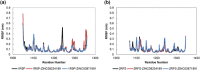
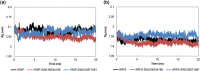
c-Met receptor tyrosine kinase is a proto-oncogene whose aberrant activation is attributed to a lower rate of survival in most cancers. Natural product-derived inhibitors known as "fourth generation inhibitors" constitute more than 60% of anticancer drugs. Furthermore, consensus docking approach has recently been introduced to augment docking accuracy and reduce false positives during a virtual screening. In order to obtain novel small-molecule Met inhibitors, consensus docking approach was performed using Autodock Vina and Autodock 4.2 to virtual screen Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target database against active and inactive conformation of c-Met kinase domain structure. Two hit molecules that were in line with drug-likeness criteria, desired docking score, and binding pose were subjected to molecular dynamics simulations to elucidate intermolecular contacts in protein-ligand complexes. Analysis of molecular dynamics simulations and molecular mechanics Poisson-Boltzmann surface area studies showed that ZINC08234189 is a plausible inhibitor for the active state of c-Met, whereas ZINC03871891 may be more effective toward active c-Met kinase domain compared to the inactive form due to higher binding energy. Our analysis showed that both the hit molecules formed hydrogen bonds with key residues of the hinge region (P1158, M1160) in the active form, which is a hallmark of kinase domain inhibitors. Considering the pivotal role of HGF/c-Met signaling in carcinogenesis, our results propose ZINC08234189 and ZINC03871891 as the therapeutic options to surmount Met-dependent cancers.
Keywords: Binding free energy; Consensus docking; Molecular dynamics simulation; Natural products; c-Met inhibitor.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

