Advances and challenges in the detection of transcriptome-wide protein-RNA interactions
- PMID: 28853213
- PMCID: PMC5739989
- DOI: 10.1002/wrna.1436
Advances and challenges in the detection of transcriptome-wide protein-RNA interactions
Abstract
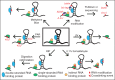
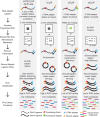
RNA binding proteins (RBPs) play key roles in determining cellular behavior by manipulating the processing of target RNAs. Robust methods are required to detect the numerous binding sites of RBPs across the transcriptome. RNA-immunoprecipitation followed by sequencing (RIP-seq) and crosslinking followed by immunoprecipitation and sequencing (CLIP-seq) are state-of-the-art methods used to identify the RNA targets and specific binding sites of RBPs. Historically, CLIP methods have been confounded with challenges such as the requirement for tens of millions of cells per experiment, low RNA yields resulting in libraries that contain a high number of polymerase chain reaction duplicated reads, and technical inconveniences such as radioactive labeling of RNAs. However, recent improvements in the recovery of bound RNAs and the efficiency of converting isolated RNAs into a library for sequencing have enhanced our ability to perform the experiment at scale, from less starting material than has previously been possible, and resulting in high quality datasets for the confident identification of protein binding sites. These, along with additional improvements to protein capture, removal of nonspecific signals, and methods to isolate noncanonical RBP targets have revolutionized the study of RNA processing regulation, and reveal a promising future for mapping the human protein-RNA regulatory network. WIREs RNA 2018, 9:e1436. doi: 10.1002/wrna.1436 This article is categorized under: RNA Interactions with Proteins and Other Molecules > Protein-RNA Recognition RNA Interactions with Proteins and Other Molecules > Protein-RNA Interactions: Functional Implications RNA Methods > RNA Analyses in Cells.
© 2017 The Authors. WIREs RNA published by Wiley Periodicals, Inc.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

