Direct Observation of Insulin Association Dynamics with Time-Resolved X-ray Scattering
- PMID: 28853898
- PMCID: PMC5804350
- DOI: 10.1021/acs.jpclett.7b01720
Direct Observation of Insulin Association Dynamics with Time-Resolved X-ray Scattering
Abstract
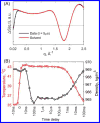
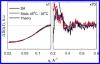
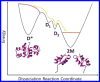
Biological functions frequently require protein-protein interactions that involve secondary and tertiary structural perturbation. Here we study protein-protein dissociation and reassociation dynamics in insulin, a model system for protein oligomerization. Insulin dimer dissociation into monomers was induced by a nanosecond temperature-jump (T-jump) of ∼8 °C in aqueous solution, and the resulting protein and solvent dynamics were tracked by time-resolved X-ray solution scattering (TRXSS) on time scales of 10 ns to 100 ms. The protein scattering signals revealed the formation of five distinguishable transient species during the association process that deviate from simple two-state kinetics. Our results show that the combination of T-jump pump coupled to TRXSS probe allows for direct tracking of structural dynamics in nonphotoactive proteins.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Nooren IMA, Thornton JM. Structural Characterisation and Functional Significance of Transient Protein-Protein Interactions. J. Mol. Biol. 2003;325(5):991–1018. - PubMed
-
- Schwarze S, Zwettler FU, Johnson CM, Neuweiler H, Boles JO. The N-Terminal Domains of Spider Silk Proteins Assemble Ultrafast and Protected from Charge Screening. Nat. Commun. 2013;4:179–182. - PubMed
-
- Sato D, Ohtomo H, Yamada Y, Hikima T, Kurobe A, Fujiwara K, Ikeguchi M. Ferritin Assembly Revisited: A Time- Resolved Small-Angle X-Ray Scattering Study. Biochemistry. 2016;55(2):287–293. - PubMed
-
- Schuck P. On the Analysis of Protein Self-Association by Sedimentation Velocity Analytical Ultracentrifugation. Anal. Biochem. 2003;320(1):104–124. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

