Regeneration Genetics
- PMID: 28853919
- PMCID: PMC5705327
- DOI: 10.1146/annurev-genet-120116-024554
Regeneration Genetics
Abstract
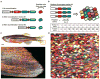
Understanding how and why animals regenerate complex tissues has the potential to transform regenerative medicine. Here we present an overview of genetic approaches that have recently been applied to dissect mechanisms of regeneration. We describe new advances that relate to central objectives of regeneration biologists researching different tissues and species, focusing mainly on vertebrates. These objectives include defining the cellular sources and key cell behaviors in regenerating tissue, elucidating molecular triggers and brakes for regeneration, and defining the earliest events that control the presence of these molecular factors.
Keywords: blastema; genetics; imaging; regeneration; salamanders; zebrafish.
Figures


References
-
- Abremski K, Hoess R. Bacteriophage P1 site-specific recombination. Purification and properties of the Cre recombinase protein. J Biol Chem. 1984;259:1509–14. - PubMed
-
- Adler PN, MacQueen M. Cell proliferation and DNA replication in the imaginal wing disc of Drosophila melanogaster. Dev Biol. 1984;103:28–37. - PubMed
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

