Evolution and functional characterization of CAZymes belonging to subfamily 10 of glycoside hydrolase family 5 (GH5_10) in two species of phytophagous beetles
- PMID: 28854242
- PMCID: PMC5576741
- DOI: 10.1371/journal.pone.0184305
Evolution and functional characterization of CAZymes belonging to subfamily 10 of glycoside hydrolase family 5 (GH5_10) in two species of phytophagous beetles
Abstract
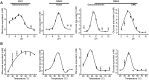
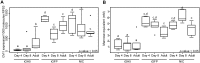
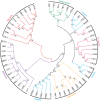
Hemicelluloses, such as xyloglucan, xylan and mannans, consist of a heterogeneous array of plant-derived polysaccharides that form the plant cell wall. These polysaccharides differ from each other in their structure and physiochemical properties, but they share a β-(1,4)-linked sugar backbone. Hemicelluloses can be hydrolyzed by plant-cell-wall-degrading enzymes (PCWDEs), which are widely distributed in phytopathogenic microbes. Recently, it has become apparent that phytophagous beetles also produce their own PCWDEs. Our previous work identified genes encoding putative mannanases belonging to the subfamily 10 of glycoside hydrolase (GH) family 5 (GH5_10) in the genomes of the leaf beetle, Gastrophysa viridula (Chrysomelidae, Chrysomelinae; one gene), and of the bean beetle, Callosobruchus maculatus (Chrysomelidae, Bruchinae; four genes). In contrast to proteins from other GH5 subfamilies, GH5_10 proteins are patchily distributed within the tree of life and have so far hardly been investigated. We addressed the following questions: Are beetle-derived GH5_10s active PCWDEs? How did they evolve? What is their physiological function? Using heterologous protein expression and enzymatic assays, we show that the G. viridula GH5_10 protein is an endo-β-1,4-mannanase. We also demonstrate that only one out of four C. maculatus GH5_10 proteins is an endo-β-1,4-mannanase, which has additional activity on carboxymethyl cellulose. Unexpectedly, another C. maculatus GH5_10 protein has evolved to use xylan instead of mannans as a substrate. RNAi experiments in G. viridula indicate (i) that the sole GH5_10 protein is responsible for breaking down mannans in the gut and (ii) that this breakdown may rather be accessory and may facilitate access to plant cell content, which is rich in nitrogen and simple sugars. Phylogenetic analyses indicate that coleopteran-derived GH5_10 proteins cluster together with Chelicerata-derived ones. Interestingly, other insect-derived GH5_10 proteins cluster elsewhere, suggesting insects have several independent evolutionary origins.
Conflict of interest statement
Figures



References
-
- Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861. doi: 10.1038/nrm1746 - DOI - PubMed
-
- Cosgrove DJ (2014) Re-constructing our models of cellulose and primary cell wall assembly. Curr Opin Plant Biol 22: 122–131. doi: 10.1016/j.pbi.2014.11.001 - DOI - PMC - PubMed
-
- Rodriguez-Gacio Mdel C, Iglesias-Fernandez R, Carbonero P, Matilla AJ (2012) Softening-up mannan-rich cell walls. J Exp Bot 63: 3976–3988. doi: 10.1093/jxb/ers096 - DOI - PubMed
-
- Meier H, Reid JSG (1982) Reserve Polysaccharides Other Than Starch in Higher Plants In: Loewus FA, Tanner W, editors. Plant Carbohydrates I. Berlin Heidelberg New York: Springer-Verlag; pp. 418–471.
-
- Buckeridge MS, dos Santos HP, Tine MAS (2000) Mobilisation of storage cell wall polysaccharides in seeds. Plant Physiol Biochem 38: 141–156.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

