Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants
- PMID: 28854567
- PMCID: PMC5737848
- DOI: 10.1093/aob/mcx079
Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants
Erratum in
-
Erratum.Ann Bot. 2017 Oct 17;120(4):619. doi: 10.1093/aob/mcx096. Ann Bot. 2017. PMID: 28961731 Free PMC article. No abstract available.
Abstract
Background: Polyploidy or whole-genome duplication is now recognized as being present in almost all lineages of higher plants, with multiple rounds of polyploidy occurring in most extant species. The ancient evolutionary events have been identified through genome sequence analysis, while recent hybridization events are found in about half of the world's crops and wild species. Building from this new paradigm for understanding plant evolution, the papers in this Special Issue address questions about polyploidy in ecology, adaptation, reproduction and speciation of wild and cultivated plants from diverse ecosystems. Other papers, including this review, consider genomic aspects of polyploidy.
Approaches: Discovery of the evolutionary consequences of new, evolutionarily recent and ancient polyploidy requires a range of approaches. Large-scale studies of both single species and whole ecosystems, with hundreds to tens of thousands of individuals, sometimes involving 'garden' or transplant experiments, are important for studying adaptation. Molecular studies of genomes are needed to measure diversity in genotypes, showing ancestors, the nature and number of polyploidy and backcross events that have occurred, and allowing analysis of gene expression and transposable element activation. Speciation events and the impact of reticulate evolution require comprehensive phylogenetic analyses and can be assisted by resynthesis of hybrids. In this Special Issue, we include studies ranging in scope from experimental and genomic, through ecological to more theoretical.
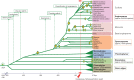
Conclusions: The success of polyploidy, displacing the diploid ancestors of almost all plants, is well illustrated by the huge angiosperm diversity that is assumed to originate from recurrent polyploidization events. Strikingly, polyploidization often occurred prior to or simultaneously with major evolutionary transitions and adaptive radiation of species, supporting the concept that polyploidy plays a predominant role in bursts of adaptive speciation. Polyploidy results in immediate genetic redundancy and represents, with the emergence of new gene functions, an important source of novelty. Along with recombination, gene mutation, transposon activity and chromosomal rearrangement, polyploidy and whole-genome duplication act as drivers of evolution and divergence in plant behaviour and gene function, enabling diversification, speciation and hence plant evolution.
Keywords: Polyploidy; adaptation; angiosperms; bryophytes; chromosomes; crops; ecology; evolution; genomics; hybrids; phylogeny; speciation; weeds; whole-genome duplication (WGD).
© The Author 2017. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For Permissions, please email: journals.permissions@oup.com
Figures

References
-
- Ågren JA. 2013. Selfish genes and plant speciation. Evolutionary Biology 40: 439–449.
-
- Ågren JA, Wright SI.. 2011. Co-evolution between transposable elements and their hosts: a major factor in genome size evolution? Chromosome Research 19: 777–786. - PubMed
-
- Alix K, Joets J, Ryder CD, et al. 2008. The CACTA transposon Bot1 played a major role in Brassica genome divergence and gene proliferation. The Plant Journal 56: 1030–1044. - PubMed
-
- Amborella Genome Project. 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

