Evolutionary Dynamics of Cryptophyte Plastid Genomes
- PMID: 28854597
- PMCID: PMC5534331
- DOI: 10.1093/gbe/evx123
Evolutionary Dynamics of Cryptophyte Plastid Genomes
Abstract
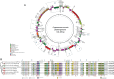
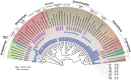
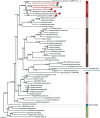
Cryptophytes are an ecologically important group of largely photosynthetic unicellular eukaryotes. This lineage is of great interest to evolutionary biologists because their plastids are of red algal secondary endosymbiotic origin and the host cell retains four different genomes (host nuclear, mitochondrial, plastid, and red algal nucleomorph). Here, we report a comparative analysis of plastid genomes from six representative cryptophyte genera. Four newly sequenced cryptophyte plastid genomes of Chroomonas mesostigmatica, Ch. placoidea, Cryptomonas curvata, and Storeatula sp. CCMP1868 share a number of features including synteny and gene content with the previously sequenced genomes of Cryptomonas paramecium, Rhodomonas salina, Teleaulax amphioxeia, and Guillardia theta. Our analysis of these plastid genomes reveals examples of gene loss and intron insertion. In particular, the chlB/chlL/chlN genes, which encode light-independent (dark active) protochlorophyllide oxidoreductase (LIPOR) proteins have undergone recent gene loss and pseudogenization in cryptophytes. Comparison of phylogenetic trees based on plastid and nuclear genome data sets show the introduction, via secondary endosymbiosis, of a red algal derived plastid in a lineage of chlorophyll-c containing algae. This event was followed by additional rounds of eukaryotic endosymbioses that spread the red lineage plastid to diverse groups such as haptophytes and stramenopiles.
Keywords: cryptophyte; horizontal gene transfer; plastid genome.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Comparative mitochondrial genomics of cryptophyte algae: gene shuffling and dynamic mobile genetic elements.BMC Genomics. 2018 Apr 20;19(1):275. doi: 10.1186/s12864-018-4626-9. BMC Genomics. 2018. PMID: 29678149 Free PMC article.
-
Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny.Mol Biol Evol. 2007 Aug;24(8):1832-42. doi: 10.1093/molbev/msm101. Epub 2007 May 23. Mol Biol Evol. 2007. PMID: 17522086
-
The Plastid Genome of the Cryptomonad Teleaulax amphioxeia.PLoS One. 2015 Jun 5;10(6):e0129284. doi: 10.1371/journal.pone.0129284. eCollection 2015. PLoS One. 2015. PMID: 26047475 Free PMC article.
-
After the primary endosymbiosis: an update on the chromalveolate hypothesis and the origins of algae with Chl c.Photosynth Res. 2011 Jan;107(1):103-15. doi: 10.1007/s11120-010-9584-2. Epub 2010 Jul 30. Photosynth Res. 2011. PMID: 20676772 Review.
-
A new scenario of plastid evolution: plastid primary endosymbiosis before the divergence of the "Plantae," emended.J Plant Res. 2005 Aug;118(4):247-55. doi: 10.1007/s10265-005-0219-1. Epub 2005 Jul 20. J Plant Res. 2005. PMID: 16032387 Review.
Cited by
-
Comparative analysis of plastome structure in Sphagnum species from China.BMC Genomics. 2025 Jul 1;26(1):585. doi: 10.1186/s12864-025-11763-y. BMC Genomics. 2025. PMID: 40597574 Free PMC article.
-
The structure of PSI-LHCI from Cyanidium caldarium provides evolutionary insights into conservation and diversity of red-lineage LHCs.Proc Natl Acad Sci U S A. 2024 Mar 12;121(11):e2319658121. doi: 10.1073/pnas.2319658121. Epub 2024 Mar 5. Proc Natl Acad Sci U S A. 2024. PMID: 38442179 Free PMC article.
-
Comparative Plastid Genomics of Non-Photosynthetic Chrysophytes: Genome Reduction and Compaction.Front Plant Sci. 2020 Sep 10;11:572703. doi: 10.3389/fpls.2020.572703. eCollection 2020. Front Plant Sci. 2020. PMID: 33013997 Free PMC article.
-
What Happened before Losses of Photosynthesis in Cryptophyte Algae?Mol Biol Evol. 2022 Feb 3;39(2):msac001. doi: 10.1093/molbev/msac001. Mol Biol Evol. 2022. PMID: 35079797 Free PMC article.
-
Comparative mitochondrial genomics of cryptophyte algae: gene shuffling and dynamic mobile genetic elements.BMC Genomics. 2018 Apr 20;19(1):275. doi: 10.1186/s12864-018-4626-9. BMC Genomics. 2018. PMID: 29678149 Free PMC article.
References
-
- Archibald JM. 2007. Nucleomorph genomes: structure, function, origin and evolution. BioEssays 29:392–402. - PubMed
-
- Archibald JM. 2009. The puzzle of plastid evolution. Curr Biol. 19:R81–R88. - PubMed
-
- Archibald JM, Keeling PJ.. 2005. On the origin and evolution of plastids In: Saap J, editor. Microbial phylogeny and evolution. New York: Oxford University Press; p. 238–260.
-
- Baurain D, et al.2010. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 27:1698–1709. - PubMed
-
- Bhattacharya D, Medlin L.. 1995. The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. J Phycol. 31:489–498.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

