Rapid Evolution of Primate Type 2 Immune Response Factors Linked to Asthma Susceptibility
- PMID: 28854632
- PMCID: PMC5569703
- DOI: 10.1093/gbe/evx120
Rapid Evolution of Primate Type 2 Immune Response Factors Linked to Asthma Susceptibility
Abstract
Host immunity pathways evolve rapidly in response to antagonism by pathogens. Microbial infections can also trigger excessive inflammation that contributes to diverse autoimmune disorders including asthma, lupus, diabetes, and arthritis. Definitive links between immune system evolution and human autoimmune disease remain unclear. Here we provide evidence that several components of the type 2 immune response pathway have been subject to recurrent positive selection in the primate lineage. Notably, substitutions in the central immune regulator IL13 correspond to a polymorphism linked to asthma susceptibility in humans. We also find evidence of accelerated amino acid substitutions as well as gene gain and loss events among eosinophil granule proteins, which act as toxic antimicrobial effectors that promote asthma pathology by damaging airway tissues. These results support the hypothesis that evolutionary conflicts with pathogens promote tradeoffs for increasingly robust immune responses during animal evolution. Our findings are also consistent with the view that natural selection has contributed to the spread of autoimmune disease alleles in humans.
Keywords: asthma genetics; eosinophil granule protein; human variation; interleukin 13; primate evolution.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

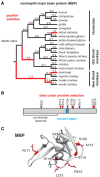

References
-
- Abascal F, Zardoya R, Posada D.. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

