Contrasting Patterns of Nucleotide Substitution Rates Provide Insight into Dynamic Evolution of Plastid and Mitochondrial Genomes of Geranium
- PMID: 28854633
- PMCID: PMC5570028
- DOI: 10.1093/gbe/evx124
Contrasting Patterns of Nucleotide Substitution Rates Provide Insight into Dynamic Evolution of Plastid and Mitochondrial Genomes of Geranium
Abstract
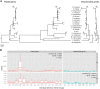
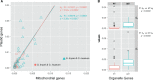
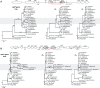
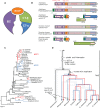
Geraniaceae have emerged as a model system for investigating the causes and consequences of variation in plastid and mitochondrial genomes. Incredible structural variation in plastid genomes (plastomes) and highly accelerated evolutionary rates have been reported in selected lineages and functional groups of genes in both plastomes and mitochondrial genomes (mitogenomes), and these phenomena have been implicated in cytonuclear incompatibility. Previous organelle genome studies have included limited sampling of Geranium, the largest genus in the family with over 400 species. This study reports on rates and patterns of nucleotide substitutions in plastomes and mitogenomes of 17 species of Geranium and representatives of other Geraniaceae. As detected across other angiosperms, substitution rates in the plastome are 3.5 times higher than the mitogenome in most Geranium. However, in the branch leading to Geranium brycei/Geranium incanum mitochondrial genes experienced significantly higher dN and dS than plastid genes, a pattern that has only been detected in one other angiosperm. Furthermore, rate accelerations differ in the two organelle genomes with plastomes having increased dN and mitogenomes with increased dS. In the Geranium phaeum/Geranium reflexum clade, duplicate copies of clpP and rpoA genes that experienced asymmetric rate divergence were detected in the single copy region of the plastome. In the case of rpoA, the branch leading to G. phaeum/G. reflexum experienced positive selection or relaxation of purifying selection. Finally, the evolution of acetyl-CoA carboxylase is unusual in Geraniaceae because it is only the second angiosperm family where both prokaryotic and eukaryotic ACCases functionally coexist in the plastid.
Keywords: accD; accelerated substitution rate; clpP; cytonuclear incompatibility; gene duplication; positive selection; rpoA.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Adams KL, Palmer JD.. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogen Evol. 29:380–395. - PubMed
-
- Aedo C, Garmenia FM, Pando F.. 1998. World checklist of Geranium L. (Geraniaceae). Anal Jard Bot Madrid. 56:211–252.
-
- Bakker FT, Breman F, Merckx V.. 2006. DNA sequence evolution in fast evolving mitochondrial DNA nad1 exons in Geraniaceae and Plantaginaceae. Taxon 55:887–896.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

