Comparative Genomics of the Dual-Obligate Symbionts from the Treehopper, Entylia carinata (Hemiptera: Membracidae), Provide Insight into the Origins and Evolution of an Ancient Symbiosis
- PMID: 28854637
- PMCID: PMC5533117
- DOI: 10.1093/gbe/evx134
Comparative Genomics of the Dual-Obligate Symbionts from the Treehopper, Entylia carinata (Hemiptera: Membracidae), Provide Insight into the Origins and Evolution of an Ancient Symbiosis
Abstract
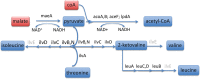
Insect species in the Auchenorrhyncha suborder (Hemiptera) maintain ancient obligate symbioses with bacteria that provide essential amino acids (EAAs) deficient in their plant-sap diets. Molecular studies have revealed that two complementary symbiont lineages, "Candidatus Sulcia muelleri" and a betaproteobacterium ("Ca. Zinderia insecticola" in spittlebugs [Cercopoidea] and "Ca. Nasuia deltocephalinicola" in leafhoppers [Cicadellidae]) may have persisted in the suborder since its origin ∼300 Ma. However, investigation of how this pair has co-evolved on a genomic level is limited to only a few host lineages. We sequenced the complete genomes of Sulcia and a betaproteobacterium from the treehopper, Entylia carinata (Membracidae: ENCA), as the first representative from this species-rich group. It also offers the opportunity to compare symbiont evolution across a major insect group, the Membracoidea (leafhoppers + treehoppers). Genomic analyses show that the betaproteobacteria in ENCA is a member of the Nasuia lineage. Both symbionts have larger genomes (Sulcia = 218 kb and Nasuia = 144 kb) than related lineages in Deltocephalinae leafhoppers, retaining genes involved in basic cellular functions and information processing. Nasuia-ENCA further exhibits few unique gene losses, suggesting that its parent lineage in the common ancestor to the Membracoidea was already highly reduced. Sulcia-ENCA has lost the abilities to synthesize menaquinone cofactor and to complete the synthesis of the branched-chain EAAs. Both capabilities are conserved in other Sulcia lineages sequenced from across the Auchenorrhyncha. Finally, metagenomic sequencing recovered the partial genome of an Arsenophonus symbiont, although it infects only 20% of individuals indicating a facultative role.
Keywords: aminoacyl tRNA synthetases; bacteria; facultative symbiont; gene loss; genome evolution; nutritional symbioses.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

