Evolution of the Largest Mammalian Genome
- PMID: 28854639
- PMCID: PMC5569995
- DOI: 10.1093/gbe/evx113
Evolution of the Largest Mammalian Genome
Abstract
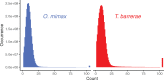
The genome of the red vizcacha rat (Rodentia, Octodontidae, Tympanoctomys barrerae) is the largest of all mammals, and about double the size of their close relative, the mountain vizcacha rat Octomys mimax, even though the lineages that gave rise to these species diverged from each other only about 5 Ma. The mechanism for this rapid genome expansion is controversial, and hypothesized to be a consequence of whole genome duplication or accumulation of repetitive elements. To test these alternative but nonexclusive hypotheses, we gathered and evaluated evidence from whole transcriptome and whole genome sequences of T. barrerae and O. mimax. We recovered support for genome expansion due to accumulation of a diverse assemblage of repetitive elements, which represent about one half and one fifth of the genomes of T. barrerae and O. mimax, respectively, but we found no strong signal of whole genome duplication. In both species, repetitive sequences were rare in transcribed regions as compared with the rest of the genome, and mostly had no close match to annotated repetitive sequences from other rodents. These findings raise new questions about the genomic dynamics of these repetitive elements, their connection to widespread chromosomal fissions that occurred in the T. barrerae ancestor, and their fitness effects-including during the evolution of hypersaline dietary tolerance in T. barrerae.
Keywords: Caviomorpha; Octodontidae; Rodentia; mammals; repetitive DNA; whole genome duplication.
©The Author(s) 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Acquaah G. 2007. Principles of plant genetics and breeding. Malden, MA: Blackwell Publishing.
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. - PubMed
-
- Andrews S, et al.2010. FastQC: A quality control tool for high throughput sequence data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Bacquet C, et al.2008. Epigenetic processes in a tetraploid mammal. Mamm Genome 19:439–447. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

