The role of poly ADP-ribosylation in the first wave of DNA damage response
- PMID: 28854736
- PMCID: PMC5737498
- DOI: 10.1093/nar/gkx565
The role of poly ADP-ribosylation in the first wave of DNA damage response
Abstract
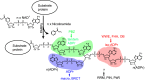
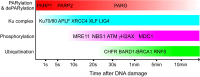
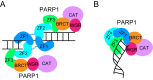
Poly ADP-ribose polymerases (PARPs) catalyze massive protein poly ADP-ribosylation (PARylation) within seconds after the induction of DNA single- or double-strand breaks. PARylation occurs at or near the sites of DNA damage and promotes the recruitment of DNA repair factors via their poly ADP-ribose (PAR) binding domains. Several novel PAR-binding domains have been recently identified. Here, we summarize these and other recent findings suggesting that PARylation may be the critical event that mediates the first wave of the DNA damage response. We also discuss the potential for functional crosstalk with other DNA damage-induced post-translational modifications.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001; 411:366–374. - PubMed
-
- Lindahl T., Barnes D.E.. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000; 65:127–133. - PubMed
-
- Valko M., Rhodes C.J., Moncol J., Izakovic M., Mazur M.. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006; 160:1–40. - PubMed
-
- Zhou B.B., Elledge S.J.. The DNA damage response: putting checkpoints in perspective. Nature. 2000; 408:433–439. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

