Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors
- PMID: 28854875
- PMCID: PMC5577687
- DOI: 10.1186/s12864-017-4087-6
Xanthomonas adaptation to common bean is associated with horizontal transfers of genes encoding TAL effectors
Abstract
Background: Common bacterial blight is a devastating bacterial disease of common bean (Phaseolus vulgaris) caused by Xanthomonas citri pv. fuscans and Xanthomonas phaseoli pv. phaseoli. These phylogenetically distant strains are able to cause similar symptoms on common bean, suggesting that they have acquired common genetic determinants of adaptation to common bean. Transcription Activator-Like (TAL) effectors are bacterial type III effectors that are able to induce the expression of host genes to promote infection or resistance. Their capacity to bind to a specific host DNA sequence suggests that they are potential candidates for host adaption.
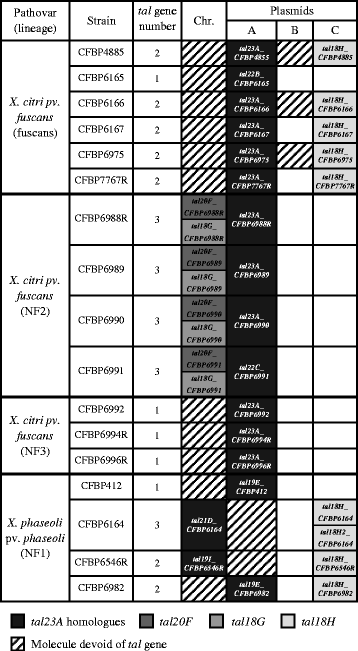
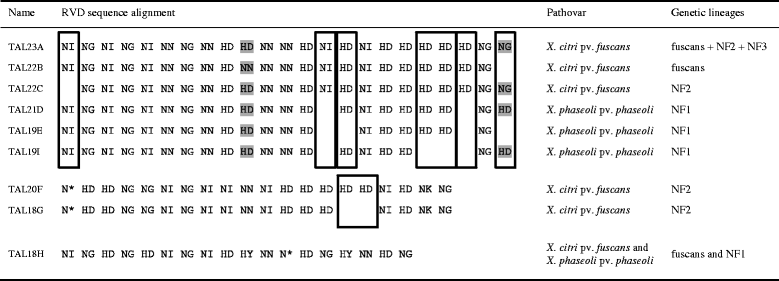
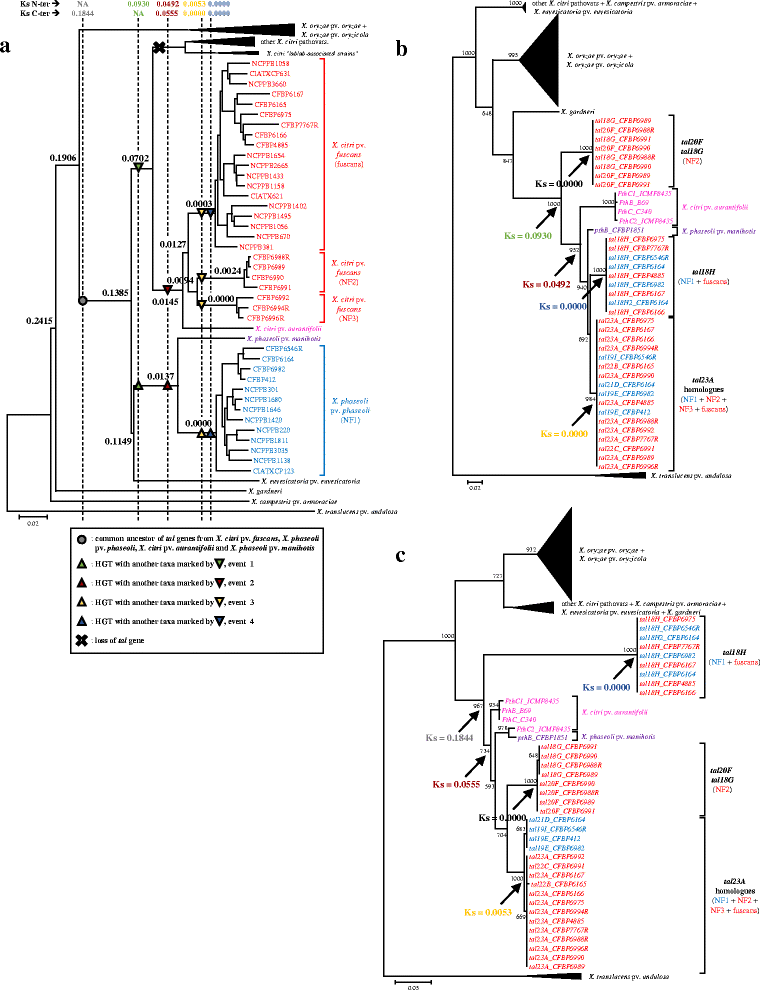
Results: To study the diversity of tal genes from Xanthomonas strains responsible for common bacterial blight of bean, whole genome sequences of 17 strains representing the diversity of X. citri pv. fuscans and X. phaseoli pv. phaseoli were obtained by single molecule real time sequencing. Analysis of these genomes revealed the existence of four tal genes named tal23A, tal20F, tal18G and tal18H, respectively. While tal20F and tal18G were chromosomic, tal23A and tal18H were carried on plasmids and shared between phylogenetically distant strains, therefore suggesting recent horizontal transfers of these genes between X. citri pv. fuscans and X. phaseoli pv. phaseoli strains. Strikingly, tal23A was present in all strains studied, suggesting that it played an important role in adaptation to common bean. In silico predictions of TAL effectors targets in the common bean genome suggested that TAL effectors shared by X. citri pv. fuscans and X. phaseoli pv. phaseoli strains target the promoters of genes of similar functions. This could be a trace of convergent evolution among TAL effectors from different phylogenetic groups, and comforts the hypothesis that TAL effectors have been implied in the adaptation to common bean.
Conclusions: Altogether, our results favour a model where plasmidic TAL effectors are able to contribute to host adaptation by being horizontally transferred between distant lineages.
Keywords: Common bean; Horizontal gene transfer; Host adaptation; TAL effectors; Xanthomonas.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

