Comparison of rumen bacterial communities in dairy herds of different production
- PMID: 28854878
- PMCID: PMC5577838
- DOI: 10.1186/s12866-017-1098-z
Comparison of rumen bacterial communities in dairy herds of different production
Abstract
Background: The purpose of this study was to compare the rumen bacterial composition in high and low yielding dairy cows within and between two dairy herds. Eighty five Holstein dairy cows in mid-lactation (79-179 days in milk) were selected from two farms: Farm 12 (M305 = 12,300 kg; n = 47; 24 primiparous cows, 23 multiparous cows) and Farm 9 (M305 = 9700 kg; n = 38; 19 primiparous cows, 19 multiparous cows). Each study cow was sampled once using the stomach tube method and processed for 16S rRNA gene amplicon sequencing using the Ion Torrent (PGM) platform.
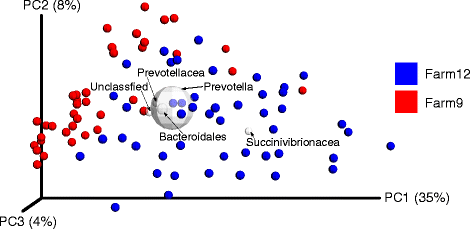
Results: Differences in bacterial communities between farms were greater (Adonis: R2 = 0.16; p < 0.001) than within farm. Five bacterial lineages, namely Prevotella (48-52%), unclassified Bacteroidales (10-12%), unclassified bacteria (5-8%), unclassified Succinivibrionaceae (1-7%) and unclassified Prevotellaceae (4-5%) were observed to differentiate the community clustering patterns among the two farms. A notable finding is the greater (p < 0.05) contribution of Succinivibrionaceae lineages in Farm 12 compared to Farm 9. Furthermore, in Farm 12, Succinivibrionaceae lineages were higher (p < 0.05) in the high yielding cows compared to the low yielding cows in both primiparous and multiparous groups. Prevotella, S24-7 and Succinivibrionaceae lineages were found in greater abundance on Farm 12 and were positively correlated with milk yield.
Conclusions: Differences in rumen bacterial populations observed between the two farms can be attributed to dietary composition, particularly differences in forage type and proportion in the diets. A combination of corn silage and alfalfa silage may have contributed to the increased proportion of Proteobacteria in Farm 12. It was concluded that Farm 12 had a greater proportion of specialist bacteria that have the potential to enhance rumen fermentative digestion of feedstuffs to support higher milk yields.
Keywords: Dairy cows; Dairy herds; Rumen microbiota.
Conflict of interest statement
Ethics approval
Dairy cows that were donors of rumen fluid were maintained according to the ethics committee and IACUC standards for the University of Pennsylvania (approval #805538).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Variability, stability, and resilience of fecal microbiota in dairy cows fed whole crop corn silage.Appl Microbiol Biotechnol. 2017 Aug;101(16):6355-6364. doi: 10.1007/s00253-017-8348-8. Epub 2017 Jun 10. Appl Microbiol Biotechnol. 2017. PMID: 28601892
-
Dried distillers grains plus solubles with corn silage or alfalfa hay as the primary forage source in dairy cow diets.J Dairy Sci. 2007 Dec;90(12):5587-99. doi: 10.3168/jds.2006-753. J Dairy Sci. 2007. PMID: 18024751
-
Effects of essential oils on digestion, ruminal fermentation, rumen microbial populations, milk production, and milk composition in dairy cows fed alfalfa silage or corn silage.J Dairy Sci. 2007 Feb;90(2):886-97. doi: 10.3168/jds.S0022-0302(07)71572-2. J Dairy Sci. 2007. PMID: 17235165
-
Symposium review: Understanding diet-microbe interactions to enhance productivity of dairy cows.J Dairy Sci. 2018 Aug;101(8):7661-7679. doi: 10.3168/jds.2017-13858. Epub 2018 May 30. J Dairy Sci. 2018. PMID: 29859694 Review.
-
Animal board invited review: The effect of diet on rumen microbial composition in dairy cows.Animal. 2024 Oct;18(10):101319. doi: 10.1016/j.animal.2024.101319. Epub 2024 Sep 2. Animal. 2024. PMID: 39305824 Review.
Cited by
-
Correlations between the Composition of the Bovine Microbiota and Vitamin B12 Abundance.mSystems. 2020 Mar 3;5(2):e00107-20. doi: 10.1128/mSystems.00107-20. mSystems. 2020. PMID: 32127420 Free PMC article.
-
Understanding the differences in rumen bacteria and their impact on dairy cows' production performance: A review.Anim Nutr. 2025 Jul 9;22:259-279. doi: 10.1016/j.aninu.2025.04.006. eCollection 2025 Sep. Anim Nutr. 2025. PMID: 40896480 Free PMC article. Review.
-
Camelina sativa L. Oil Mitigates Enteric in vitro Methane Production, Modulates Ruminal Fermentation, and Ruminal Bacterial Diversity in Buffaloes.Front Vet Sci. 2020 Aug 26;7:550. doi: 10.3389/fvets.2020.00550. eCollection 2020. Front Vet Sci. 2020. PMID: 33005640 Free PMC article.
-
A Cross-Sectional Study of Dairy Cattle Metagenomes Reveals Increased Antimicrobial Resistance in Animals Farmed in a Heavy Metal Contaminated Environment.Front Microbiol. 2020 Nov 16;11:590325. doi: 10.3389/fmicb.2020.590325. eCollection 2020. Front Microbiol. 2020. PMID: 33304338 Free PMC article.
-
The Response of Fecal Microbiota and Host Metabolome in Dairy Cows Following Rumen Fluid Transplantation.Front Microbiol. 2022 Jul 13;13:940158. doi: 10.3389/fmicb.2022.940158. eCollection 2022. Front Microbiol. 2022. PMID: 35923396 Free PMC article.
References
-
- Bergman E. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70(2):567–590. - PubMed
-
- Dewhurst R, Davies D, Merry R. Microbial protein supply from the rumen. Anim Feed Sci Technol. 2000;85(1):1–21. doi: 10.1016/S0377-8401(00)00139-5. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

