An Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function
- PMID: 28855300
- PMCID: PMC5623859
- DOI: 10.15252/emmm.201707672
An Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function
Abstract
Sequence variations occurring in the gene encoding the triggering receptor expressed on myeloid cells 2 (TREM2) support an essential function of microglia and innate immunity in the pathogenesis of Alzheimer's disease (AD) and other neurodegenerative disorders. TREM2 matures within the secretory pathway, and its ectodomain is shed on the plasma membrane. Missense mutations in the immunoglobulin (Ig)-like domain such as p.T66M and p.Y38C retain TREM2 within the endoplasmic reticulum and reduce shedding as well as TREM2-dependent phagocytosis. Using mass spectrometry, we have now determined the cleavage site of TREM2. TREM2 is shed by proteases of the ADAM (a disintegrin and metalloproteinase domain containing protein) family C-terminal to histidine 157, a position where an AD-associated coding variant has been discovered (p.H157Y) in the Han Chinese population. Opposite to the characterized mutations within the Ig-like domain, such as p.T66M and p.Y38C, the p.H157Y variant within the stalk region leads to enhanced shedding of TREM2. Elevated ectodomain shedding reduces cell surface full-length TREM2 and lowers TREM2-dependent phagocytosis. Therefore, two seemingly opposite cellular effects of TREM2 variants, namely reduced versus enhanced shedding, result in similar phenotypic outcomes by reducing cell surface TREM2.
Keywords: Alzheimer's disease; TREM2; neurodegeneration; phagocytosis; regulated intramembrane proteolysis.
© 2017 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
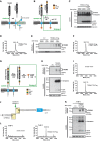
Regulated intramembrane proteolysis of TREM2. ADAM10 initiates proteolytic processing of TREM2 by liberating its ectodomain (sTREM2). Subsequent processing of the membrane retained C‐terminal fragment (CTF) by γ‐secretase within the transmembrane domain (TM) releases the intracellular domain (ICD) into the cytosol. A short p3‐like peptide (Haass et al, 1993) may be secreted.
Outline of strategy I for mass spectrometric (MS) determination of the N‐terminal end of the TREM2 CTF enriched upon γ‐secretase inhibition using DAPT.
Western blot analysis of TREM2 stably expressed in HEK293 Flp‐In cells upon γ‐secretase inhibition using DAPT. Application of DAPT leads to a robust accumulation of the TREM2 CTF under constitutive conditions as well as upon phorbol 12‐myristate 13‐acetate (PMA) mediated stimulation of TREM2 ectodomain shedding. The TREM2 9D11 antibody raised against the TREM2 C‐terminus was used to detect TREM2, and calnexin levels were analyzed as a loading control. Here, as in all other Western blots in Figs 1 and 2, the two lanes represent samples from two separate wells seeded at the same time.
MALDI‐TOF MS determination of the ectodomain cleavage site by immunoprecipitation of TREM2 CTF. The peak at 8,841.48 Da corresponds to a single cleavage site between histidine 157 and serine 158. Very minor additional peaks may represent cellular degradation products, as the N‐terminal counterpart cannot be observed (see Fig 1I).
Stimulation of ectodomain shedding by PMA leads to strong increases in sTREM2 (anti‐HA, upper panel) and sAPPα (2D8, lower panel). DMSO served as a vehicle control. EV, empty vector control.
MALDI‐TOF MS determination of the ectodomain cleavage site upon ADAM17 stimulation using PMA. TREM2 CTFs were enriched by γ‐secretase inhibition using DAPT (Fig 1C). The peak at 8,837.09 Da corresponds to a single cleavage site between histidine 157 and serine 158.
Outline of strategy II for MS determination of the C‐terminal end of sTREM2 generated by ectodomain shedding.
Western blot analysis of full‐length TREM2‐TEVFlag (middle panel) and ectodomain shedding (upper panel) upon transient transfection of HEK293 Flp‐In cells. EV, empty vector control.
MALDI‐TOF MS determination of the ectodomain cleavage site following the strategy outlined in Fig 1G. The peak at a mass of 3,947.78 Da corresponds to cleavage C‐terminal of histidine 157 and is absent in cells transfected with an empty expression vector.
TREM2 is shed predominantly C‐terminal to histidine 157 as observed in two different cell types as well as under constitutive and PMA‐stimulated conditions.
Western blot analysis of full‐length TREM2‐TEVFlag (middle panel) and ectodomain shedding (upper panel) upon transient transfection of THP‐1 monocytes.
MALDI‐TOF MS determination of the ectodomain cleavage site in THP‐1 monocytes following the strategy outlined in Fig 1G. The peak at a mass of 3,943.80 Da corresponds to cleavage C‐terminal of histidine 157 and is absent in cells transfected with an empty expression vector.

The TREM2 variant p.H157Y is located at the P1 site where shedding occurs via proteases of the ADAM family. In addition, the location of the p.T66M mutation within the Ig‐like V‐type domain is indicated.
Increased shedding of TREM2 p.H157Y. Western blot analysis of conditioned media of HEK293 Flp‐In cells stably transfected with cDNAs encoding HA and Flag‐tagged wt TREM2 or TREM2 p.T66M and p.H157Y. Anti‐HA antibody was used for detecting TREM2, and sAPPα levels were analyzed as a loading control. EV, empty vector control.
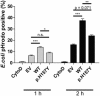
ELISA‐mediated quantitation of sTREM2 (Suarez‐Calvet et al, 2016a) in conditioned media from HEK293 cells stably expressing wt TREM2 or TREM2 p.T66M and p.H157Y. Error bars indicate SEM of seven independent experiments. One‐way ANOVA (with Dunnett's post hoc test against wt) was used for statistical analysis; wt versus EV: ***P < 0.0001; wt versus p.T66M: ***P < 0.0001; wt versus p.H157Y: **P = 0.002.
MALDI‐TOF MS analysis of the CTF derived from shedding of TREM2 p.H157Y identifies a single peak at a mass of 8,832.76 Da corresponding to a single cleavage site between tyrosine 157 and serine 158.
TREM2 p.H157Y is shed predominantly C‐terminal to tyrosine 157.
Western blot analysis of TREM2 ectodomain shedding in the presence and absence of ADAM protease inhibitors. In line with previous findings (Kleinberger et al, 2014), the broad ADAM inhibitor (GM6001; left panel) and the more selective ADAM10 inhibitor (GI254023X; right panel) reduce shedding of wt TREM2, TREM2 p.H157Y as well as sAPPα.
Western blot analysis of membrane fractions of HEK293 Flp‐In cells stably expressing wt TREM2, TREM2 p.T66M, or TREM2 p.H157Y. The TREM2 9D11 antibody raised against the TREM2 C‐terminus was used to detect TREM2, and calnexin levels were analyzed as a loading control.
Western blot analysis of membrane fractions derived from cells stably expressing wt TREM2, TREM2 p.T66M, and TREM2 p.H157Y. γ‐Secretase inhibition allows robust accumulation of the TREM2 CTF after shedding of TREM2 p.H157Y. Calnexin levels were analyzed as a loading control.
Western blot analysis of membrane fractions derived from cells stably expressing wt TREM2, TREM2 p.T66M, and TREM2 p.H157Y together with or without transiently expressed human DAP12. Note that co‐expression of human DAP12 allows the recovery of large amounts of the TREM2 p.H157Y CTF. Calnexin levels were analyzed as a loading control.
Western blot analysis of cell surface biotinylated TREM2 reveals dramatically reduced levels of surface‐exposed TREM2 p.H157Y, whereas robust amounts of cell surface wt TREM2 are detected. The anti‐HA antibody was used to detect TREM2.
References
-
- Black RA (2002) Tumor necrosis factor‐alpha converting enzyme. Int J Biochem Cell Biol 34: 1–5 - PubMed
-
- Borroni B, Ferrari F, Galimberti D, Nacmias B, Barone C, Bagnoli S, Fenoglio C, Piaceri I, Archetti S, Bonvicini C et al (2014) Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol Aging 35: 934 e937–910 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

