Methylation-dependent DNA discrimination in natural transformation of Campylobacter jejuni
- PMID: 28855338
- PMCID: PMC5617262
- DOI: 10.1073/pnas.1703331114
Methylation-dependent DNA discrimination in natural transformation of Campylobacter jejuni
Abstract
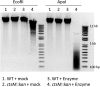
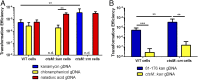
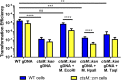
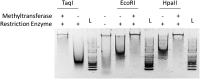
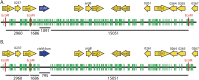
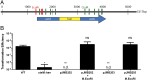
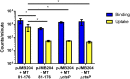
Campylobacter jejuni, a leading cause of bacterial gastroenteritis, is naturally competent. Like many competent organisms, C. jejuni restricts the DNA that can be used for transformation to minimize undesirable changes in the chromosome. Although C. jejuni can be transformed by C. jejuni-derived DNA, it is poorly transformed by the same DNA propagated in Escherichia coli or produced with PCR. Our work indicates that methylation plays an important role in marking DNA for transformation. We have identified a highly conserved DNA methyltransferase, which we term Campylobacter transformation system methyltransferase (ctsM), which methylates an overrepresented 6-bp sequence in the chromosome. DNA derived from a ctsM mutant transforms C. jejuni significantly less well than DNA derived from ctsM+ (parental) cells. The ctsM mutation itself does not affect transformation efficiency when parental DNA is used, suggesting that CtsM is important for marking transforming DNA, but not for transformation itself. The mutant has no growth defect, arguing against ongoing restriction of its own DNA. We further show that E. coli plasmid and PCR-derived DNA can efficiently transform C. jejuni when only a subset of the CtsM sites are methylated in vitro. A single methylation event 1 kb upstream of the DNA involved in homologous recombination is sufficient to transform C. jejuni, whereas otherwise identical unmethylated DNA is not. Methylation influences DNA uptake, with a slight effect also seen on DNA binding. This mechanism of DNA discrimination in C. jejuni is distinct from the DNA discrimination described in other competent bacteria.
Keywords: Campylobacter jejuni; DNA discrimination; DNA methyltransferase; competence; natural transformation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Johnsborg O, Eldholm V, Håvarstein LS. Natural genetic transformation: Prevalence, mechanisms and function. Res Microbiol. 2007;158:767–778. - PubMed
-
- Young KT, Davis LM, Dirita VJ. Campylobacter jejuni: Molecular biology and pathogenesis. Nat Rev Microbiol. 2007;5:665–679. - PubMed
-
- de Boer P, et al. Generation of Campylobacter jejuni genetic diversity in vivo. Mol Microbiol. 2002;44:351–359. - PubMed
-
- Wassenaar TM, Fry BN, van der Zeijst BA. Genetic manipulation of Campylobacter: Evaluation of natural transformation and electro-transformation. Gene. 1993;132:131–135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

