Nucleolar expansion and elevated protein translation in premature aging
- PMID: 28855503
- PMCID: PMC5577202
- DOI: 10.1038/s41467-017-00322-z
Nucleolar expansion and elevated protein translation in premature aging
Abstract
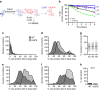
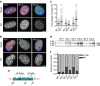
Premature aging disorders provide an opportunity to study the mechanisms that drive aging. In Hutchinson-Gilford progeria syndrome (HGPS), a mutant form of the nuclear scaffold protein lamin A distorts nuclei and sequesters nuclear proteins. We sought to investigate protein homeostasis in this disease. Here, we report a widespread increase in protein turnover in HGPS-derived cells compared to normal cells. We determine that global protein synthesis is elevated as a consequence of activated nucleoli and enhanced ribosome biogenesis in HGPS-derived fibroblasts. Depleting normal lamin A or inducing mutant lamin A expression are each sufficient to drive nucleolar expansion. We further show that nucleolar size correlates with donor age in primary fibroblasts derived from healthy individuals and that ribosomal RNA production increases with age, indicating that nucleolar size and activity can serve as aging biomarkers. While limiting ribosome biogenesis extends lifespan in several systems, we show that increased ribosome biogenesis and activity are a hallmark of premature aging.HGPS is a premature aging disease caused by mutations in the nuclear protein lamin A. Here, the authors show that cells from patients with HGPS have expanded nucleoli and increased protein synthesis, and report that nucleoli also expand as aging progresses in cells derived from healthy individuals.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Comment in
-
Ageing: Live longer with small nucleoli.Nat Rev Mol Cell Biol. 2017 Nov;18(11):651. doi: 10.1038/nrm.2017.100. Epub 2017 Sep 27. Nat Rev Mol Cell Biol. 2017. PMID: 28951566 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

