Different Adjuvants Induce Common Innate Pathways That Are Associated with Enhanced Adaptive Responses against a Model Antigen in Humans
- PMID: 28855902
- PMCID: PMC5557780
- DOI: 10.3389/fimmu.2017.00943
Different Adjuvants Induce Common Innate Pathways That Are Associated with Enhanced Adaptive Responses against a Model Antigen in Humans
Abstract
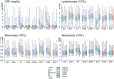
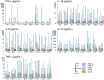
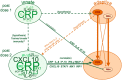
To elucidate the role of innate responses in vaccine immunogenicity, we compared early responses to hepatitis B virus (HBV) surface antigen (HBsAg) combined with different Adjuvant Systems (AS) in healthy HBV-naïve adults, and included these parameters in multi-parametric models of adaptive responses. A total of 291 participants aged 18-45 years were randomized 1:1:1:1:1 to receive HBsAg with AS01B, AS01E, AS03, AS04, or Alum/Al(OH)3 at days 0 and 30 (ClinicalTrials.gov: NCT00805389). Blood protein, cellular, and mRNA innate responses were assessed at early time-points and up to 7 days after vaccination, and used with reactogenicity symptoms in linear regression analyses evaluating their correlation with HBs-specific CD4+ T-cell and antibody responses at day 44. All AS induced transient innate responses, including interleukin (IL)-6 and C-reactive protein (CRP), mostly peaking at 24 h post-vaccination and subsiding to baseline within 1-3 days. After the second but not the first injection, median interferon (IFN)-γ levels were increased in the AS01B group, and IFN-γ-inducible protein-10 levels and IFN-inducible genes upregulated in the AS01 and AS03 groups. No distinct marker or signature was specific to one particular AS. Innate profiles were comparable between AS01B, AS01E, and AS03 groups, and between AS04 and Alum groups. AS group rankings within adaptive and innate response levels and reactogenicity prevalence were similar (AS01B ≥ AS01E > AS03 > AS04 > Alum), suggesting an association between magnitudes of inflammatory and vaccine responses. Modeling revealed associations between adaptive responses and specific traits of the innate response post-dose 2 (activation of the IFN-signaling pathway, CRP and IL-6 responses). In conclusion, the ability of AS01 and AS03 to enhance adaptive responses to co-administered HBsAg is likely linked to their capacity to activate innate immunity, particularly the IFN-signaling pathway.
Keywords: AS01; AS03; AS04; adaptive immune response; innate immune response; interferon-γ; vaccine adjuvants.
Figures




References
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

