Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov
- PMID: 28856455
- PMCID: PMC5862389
- DOI: 10.1007/s10482-017-0926-3
Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov
Abstract
The genus Elizabethkingia is genetically heterogeneous, and the phenotypic similarities between recognized species pose challenges in correct identification of clinically derived isolates. In addition to the type species Elizabethkingia meningoseptica, and more recently proposed Elizabethkingia miricola, Elizabethkingia anophelis and Elizabethkingia endophytica, four genomospecies have long been recognized. By comparing historic DNA-DNA hybridization results with whole genome sequences, optical maps, and MALDI-TOF mass spectra on a large and diverse set of strains, we propose a comprehensive taxonomic revision of this genus. Genomospecies 1 and 2 contain the type strains E. anophelis and E. miricola, respectively. Genomospecies 3 and 4 are herein proposed as novel species named as Elizabethkingia bruuniana sp. nov. (type strain, G0146T = DSM 2975T = CCUG 69503T = CIP 111191T) and Elizabethkingia ursingii sp. nov. (type strain, G4122T = DSM 2974T = CCUG 69496T = CIP 111192T), respectively. Finally, the new species Elizabethkingia occulta sp. nov. (type strain G4070T = DSM 2976T = CCUG 69505T = CIP 111193T), is proposed.
Keywords: AAI; ANI; Elizabethkingia; MALDI-TOF; SNPs; Taxonomy.
Conflict of interest statement
Figures

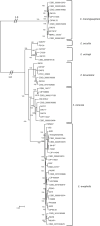
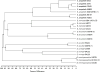
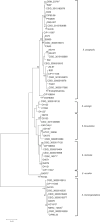
References
-
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
-
- Bloch KC, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Report of 6 cases and literature review. Medicine. 1997;76(1):30–41. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

