Elongation of very Long-Chain (>C24) Fatty Acids in Clarias gariepinus: Cloning, Functional Characterization and Tissue Expression of elovl4 Elongases
- PMID: 28856549
- PMCID: PMC5613102
- DOI: 10.1007/s11745-017-4289-3
Elongation of very Long-Chain (>C24) Fatty Acids in Clarias gariepinus: Cloning, Functional Characterization and Tissue Expression of elovl4 Elongases
Abstract
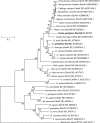
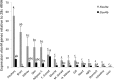
Elongation of very long-chain fatty acid 4 (Elovl4) proteins participate in the biosynthesis of very long-chain (>C24) saturated and polyunsaturated fatty acids (FA). Previous studies have shown that fish possess two different forms of Elovl4, termed Elovl4a and Elovl4b. The present study aimed to characterize both molecularly and functionally two elovl4 cDNA from the African catfish Clarias gariepinus. The results confirmed that C. gariepinus possessed two elovl4-like elongases with high homology to two previously characterized Elovl4 from Danio rerio, and thus they were termed accordingly as Elovl4a and Elovl4b. The C. gariepinus Elovl4a and Elovl4b have open reading frames (ORF) of 945 and 915 base pairs, respectively, encoding putative proteins of 314 and 304 amino acids, respectively. Functional characterization in yeast showed both Elovl4 enzymes have activity towards all the PUFA substrates assayed (18:4n-3, 18:3n-6, 20:5n-3, 20:4n-6, 22:5n-3, 22:4n-6 and 22:6n-3), producing elongated products of up to C36. Moreover, the C. gariepinus Elovl4a and Elovl4b were able to elongate very long-chain saturated FA (VLC-SFA) as denoted by increased levels of 28:0 and longer FA in yeast transformed with elovl4 ORF compared to control yeast. These results confirmed that C. gariepinus Elovl4 play important roles in the biosynthesis of very long-chain FA. Tissue distribution analysis of elovl4 mRNAs showed both genes were widely expressed in all tissues analyzed, with high expression of elovl4a in pituitary and brain, whereas female gonad and pituitary had the highest expression levels for elovl4b.
Keywords: Biosynthesis; Clarias gariepinus; Essential fatty acids; Very long-chain fatty acids; elovl4.
Figures


References
-
- Bell MV, Tocher DR. Biosynthesis of polyunsaturated fatty acids in aquatic ecosystems: general pathways and new directions. In: Arts MT, Brett M, Kainz M, editors. Lipids in aquatic ecosystems. New York: Springer; 2009. pp. 211–236.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

