Large-scale deployment of a rice 6 K SNP array for genetics and breeding applications
- PMID: 28856618
- PMCID: PMC5577349
- DOI: 10.1186/s12284-017-0181-2
Large-scale deployment of a rice 6 K SNP array for genetics and breeding applications
Abstract
Background: Fixed arrays of single nucleotide polymorphism (SNP) markers have advantages over reduced representation sequencing in their ease of data analysis, consistently higher call rates, and rapid turnaround times. A 6 K SNP array represents a cost-benefit "sweet spot" for routine genetics and breeding applications in rice. Selection of informative SNPs across species and subpopulations during chip design is essential to obtain useful polymorphism rates for target germplasm groups. This paper summarizes results from large-scale deployment of an Illumina 6 K SNP array for rice.
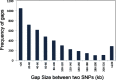
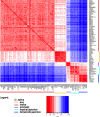
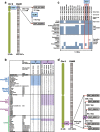
Results: Design of the Illumina Infinium 6 K SNP chip for rice, referred to as the Cornell_6K_Array_Infinium_Rice (C6AIR), includes 4429 SNPs from re-sequencing data and 1571 SNP markers from previous BeadXpress 384-SNP sets, selected based on polymorphism rate and allele frequency within and between target germplasm groups. Of the 6000 attempted bead types, 5274 passed Illumina's production quality control. The C6AIR was widely deployed at the International Rice Research Institute (IRRI) for genetic diversity analysis, QTL mapping, and tracking introgressions and was intensively used at Cornell University for QTL analysis and developing libraries of interspecific chromosome segment substitution lines (CSSLs) between O. sativa and diverse accessions of O. rufipogon or O. meridionalis. Collectively, the array was used to genotype over 40,000 rice samples. A set of 4606 SNP markers was used to provide high quality data for O. sativa germplasm, while a slightly expanded set of 4940 SNPs was used for O. sativa X O. rufipogon populations. Biparental polymorphism rates were generally between 1900 and 2500 well-distributed SNP markers for indica x japonica or interspecific populations and between 1300 and 1500 markers for crosses within indica, while polymorphism rates were lower for pairwise crosses within U.S. tropical japonica germplasm. Recently, a second-generation array containing ~7000 SNP markers, referred to as the C7AIR, was designed by removing poor-performing SNPs from the C6AIR and adding markers selected to increase the utility of the array for elite tropical japonica material.
Conclusions: The C6AIR has been successfully used to generate rapid and high-quality genotype data for diverse genetics and breeding applications in rice, and provides the basis for an optimized design in the C7AIR.
Keywords: CSSL development; High-throughput genotyping; O. rufipogon; Oryza sativa; Rice diversity; SNP fingerprinting; Single nucleotide polymorphism (SNP).
Conflict of interest statement
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
An improved 7K SNP array, the C7AIR, provides a wealth of validated SNP markers for rice breeding and genetics studies.PLoS One. 2020 May 14;15(5):e0232479. doi: 10.1371/journal.pone.0232479. eCollection 2020. PLoS One. 2020. PMID: 32407369 Free PMC article.
-
1k-RiCA (1K-Rice Custom Amplicon) a novel genotyping amplicon-based SNP assay for genetics and breeding applications in rice.Rice (N Y). 2019 Jul 26;12(1):55. doi: 10.1186/s12284-019-0311-0. Rice (N Y). 2019. PMID: 31350673 Free PMC article.
-
Development and characterization of chromosome segment substitution lines derived from Oryza rufipogon in the genetic background of O. sativa spp. indica cultivar 9311.BMC Genomics. 2016 Aug 9;17:580. doi: 10.1186/s12864-016-2987-5. BMC Genomics. 2016. PMID: 27507407 Free PMC article.
-
[Single nucleotide polymorphism (SNP) and its application in rice].Yi Chuan. 2006 Jun;28(6):737-44. Yi Chuan. 2006. PMID: 16818440 Review. Chinese.
-
Development and Applications of a High Throughput Genotyping Tool for Polyploid Crops: Single Nucleotide Polymorphism (SNP) Array.Front Plant Sci. 2018 Feb 6;9:104. doi: 10.3389/fpls.2018.00104. eCollection 2018. Front Plant Sci. 2018. PMID: 29467780 Free PMC article. Review.
Cited by
-
The Ratio of A400/A1800 Mapping Identifies Chromosomal Regions Containing Known Photoprotection Recovery-Related Genes in Rice.Rice (N Y). 2024 Sep 16;17(1):62. doi: 10.1186/s12284-024-00739-3. Rice (N Y). 2024. PMID: 39285086 Free PMC article.
-
Mapping QTLs for Reproductive Stage Salinity Tolerance in Rice Using a Cross between Hasawi and BRRI dhan28.Int J Mol Sci. 2022 Sep 27;23(19):11376. doi: 10.3390/ijms231911376. Int J Mol Sci. 2022. PMID: 36232678 Free PMC article.
-
In Silico Identification of QTL-Based Polymorphic Genes as Salt-Responsive Potential Candidates through Mapping with Two Reference Genomes in Rice.Plants (Basel). 2020 Feb 11;9(2):233. doi: 10.3390/plants9020233. Plants (Basel). 2020. PMID: 32054112 Free PMC article.
-
Genome-Wide Association Mapping to Identify Genetic Loci for Cold Tolerance and Cold Recovery During Germination in Rice.Front Genet. 2020 Feb 21;11:22. doi: 10.3389/fgene.2020.00022. eCollection 2020. Front Genet. 2020. PMID: 32153631 Free PMC article.
-
Development of GBTS and KASP Panels for Genetic Diversity, Population Structure, and Fingerprinting of a Large Collection of Broccoli (Brassica oleracea L. var. italica) in China.Front Plant Sci. 2021 Jun 4;12:655254. doi: 10.3389/fpls.2021.655254. eCollection 2021. Front Plant Sci. 2021. PMID: 34149754 Free PMC article.
References
-
- Chen S, Huang Z, Zeng L, Yang J, Liu Q, Zhu X. High-resolution mapping and gene prediction of Xanthomonas Oryzae pv. Oryzae resistance gene Xa7. Mol Breeding. 2008;22:433–441. doi: 10.1007/s11032-008-9187-1. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

