The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens
- PMID: 28856737
- PMCID: PMC6026851
- DOI: 10.1111/imr.12563
The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens
Abstract
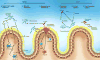
The human gastrointestinal tract hosts a diverse network of microorganisms, collectively known as the microbiota that plays an important role in health and disease. For instance, the intestinal microbiota can prevent invading microbes from colonizing the gastrointestinal tract, a phenomenon known as colonization resistance. Perturbations to the microbiota, such as antibiotic administration, can alter microbial composition and result in the loss of colonization resistance. Consequently, the host may be rendered susceptible to colonization by a pathogen. This is a particularly relevant concern in the hospital setting, where antibiotic use and antibiotic-resistant pathogen exposure are more frequent. Many nosocomial infections arise from gastrointestinal colonization. Due to their resistance to antibiotics, treatment is often very challenging. However, recent studies have demonstrated that manipulating the commensal microbiota can prevent and treat various infections in the intestine. In this review, we discuss the members of the microbiota, as well as the mechanisms, that govern colonization resistance against specific pathogens. We also review the effects of antibiotics on the microbiota, as well as the unique epidemiology of immunocompromised patients that renders them a particularly high-risk population to intestinal nosocomial infections.
Keywords: antibiotic; colonization resistance; gut; infection; microbiota.
© 2017 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

